Mercury »
PDB 1czs-1g4o »
1ej0 »
Mercury in PDB 1ej0: Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative
Protein crystallography data
The structure of Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative, PDB code: 1ej0
was solved by
H.Bugl,
E.B.Fauman,
B.L.Staker,
F.Zheng,
S.R.Kushner,
M.A.Saper,
J.C.A.Bardwell,
U.Jakob,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 17.13 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 36.579, 65.872, 72.829, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.8 / 23.2 |
Mercury Binding Sites:
The binding sites of Mercury atom in the Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative
(pdb code 1ej0). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 2 binding sites of Mercury where determined in the Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative, PDB code: 1ej0:
Jump to Mercury binding site number: 1; 2;
In total 2 binding sites of Mercury where determined in the Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative, PDB code: 1ej0:
Jump to Mercury binding site number: 1; 2;
Mercury binding site 1 out of 2 in 1ej0
Go back to
Mercury binding site 1 out
of 2 in the Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative
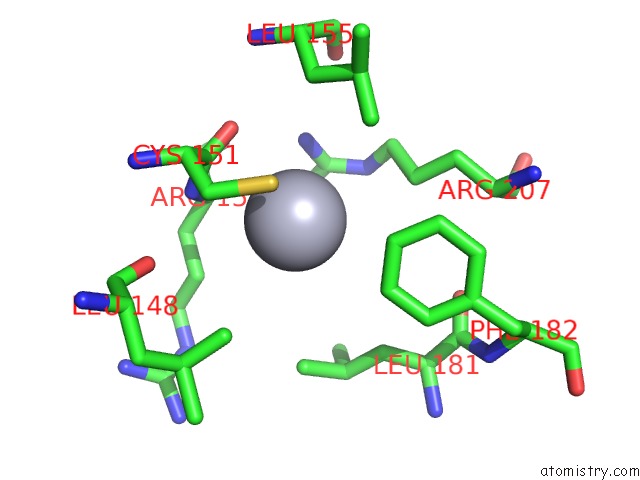
Mono view
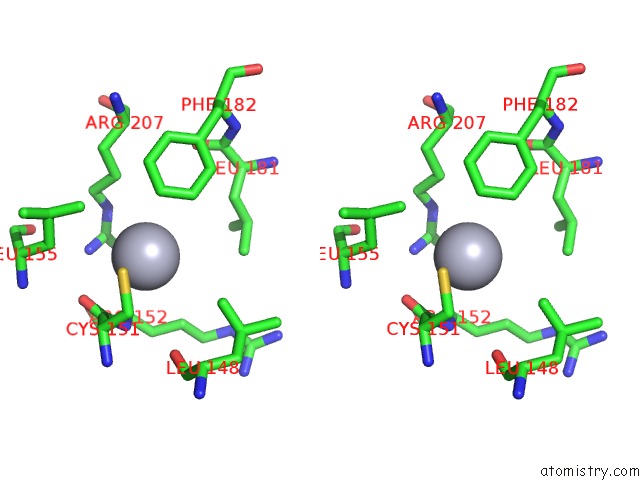
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative within 5.0Å range:
|
Mercury binding site 2 out of 2 in 1ej0
Go back to
Mercury binding site 2 out
of 2 in the Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative
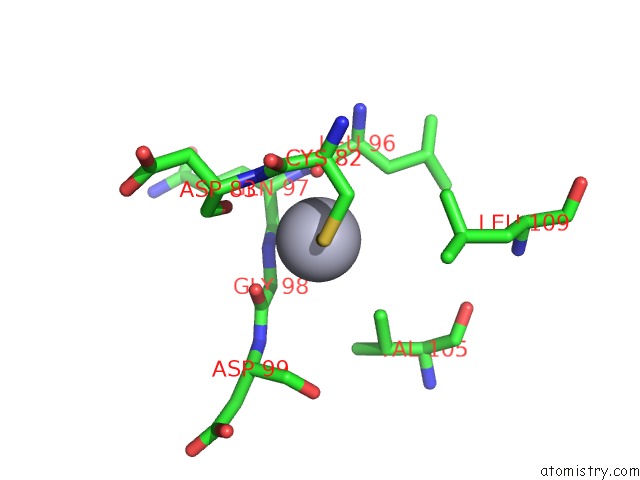
Mono view
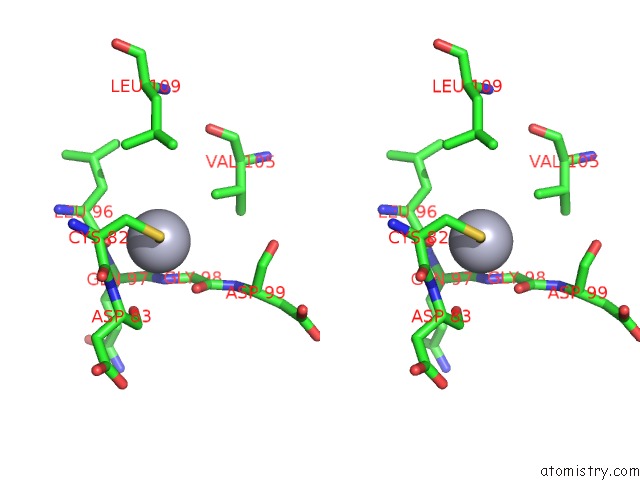
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Ftsj Rna Methyltransferase Complexed with S-Adenosylmethionine, Mercury Derivative within 5.0Å range:
|
Reference:
H.Bugl,
E.B.Fauman,
B.L.Staker,
F.Zheng,
S.R.Kushner,
M.A.Saper,
J.C.Bardwell,
U.Jakob.
Rna Methylation Under Heat Shock Control. Mol.Cell V. 6 349 2000.
ISSN: ISSN 1097-2765
PubMed: 10983982
DOI: 10.1016/S1097-2765(00)00035-6
Page generated: Sat Aug 10 23:39:53 2024
ISSN: ISSN 1097-2765
PubMed: 10983982
DOI: 10.1016/S1097-2765(00)00035-6
Last articles
Cl in 7Z7BCl in 7Z5N
Cl in 7Z7F
Cl in 7Z70
Cl in 7Z6Z
Cl in 7Z6B
Cl in 7Z6F
Cl in 7Z5M
Cl in 7Z54
Cl in 7Z50