Mercury »
PDB 1g52-1irk »
1gze »
Mercury in PDB 1gze: Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant)
Protein crystallography data
The structure of Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant), PDB code: 1gze
was solved by
J.Menetrey,
G.Flatau,
E.A.Stura,
J.B.Charbonnier,
F.Gas,
J.M.Teulon,
M.H.Le Du,
P.Boquet,
A.Menez,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 18.12 / 2.7 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 108.997, 75.596, 123.465, 90.00, 102.37, 90.00 |
| R / Rfree (%) | 24 / 29 |
Mercury Binding Sites:
The binding sites of Mercury atom in the Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant)
(pdb code 1gze). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 4 binding sites of Mercury where determined in the Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant), PDB code: 1gze:
Jump to Mercury binding site number: 1; 2; 3; 4;
In total 4 binding sites of Mercury where determined in the Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant), PDB code: 1gze:
Jump to Mercury binding site number: 1; 2; 3; 4;
Mercury binding site 1 out of 4 in 1gze
Go back to
Mercury binding site 1 out
of 4 in the Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant)
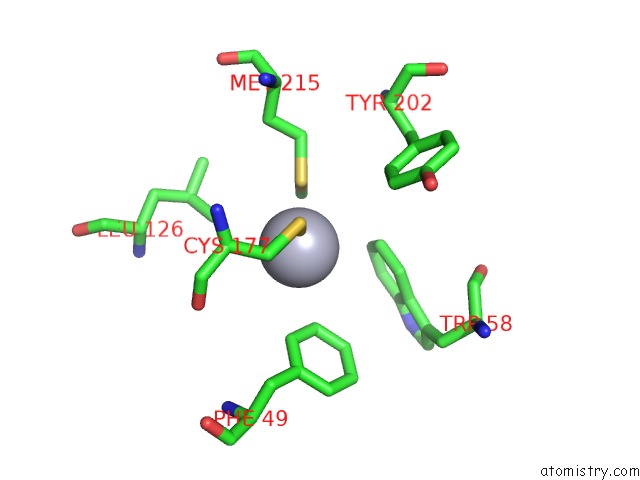
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant) within 5.0Å range:
|
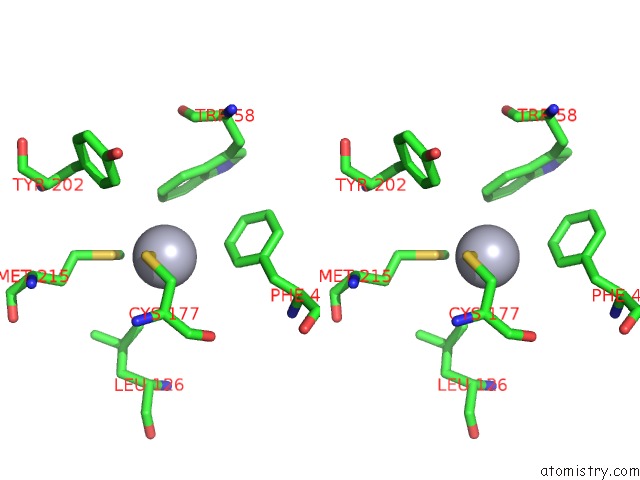
Mercury binding site 2 out of 4 in 1gze
Go back to
Mercury binding site 2 out
of 4 in the Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant)
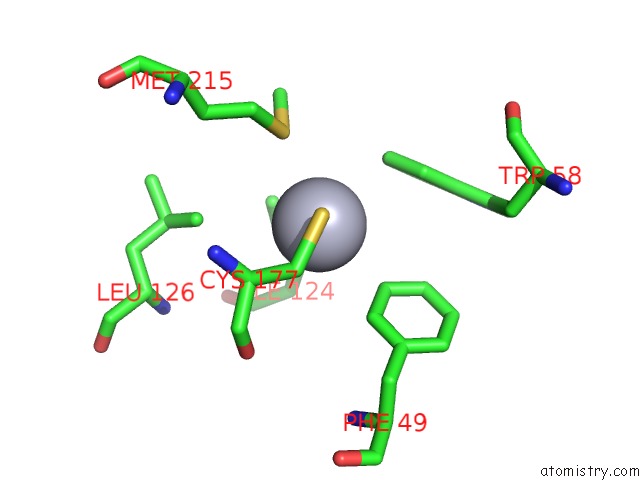
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant) within 5.0Å range:
|
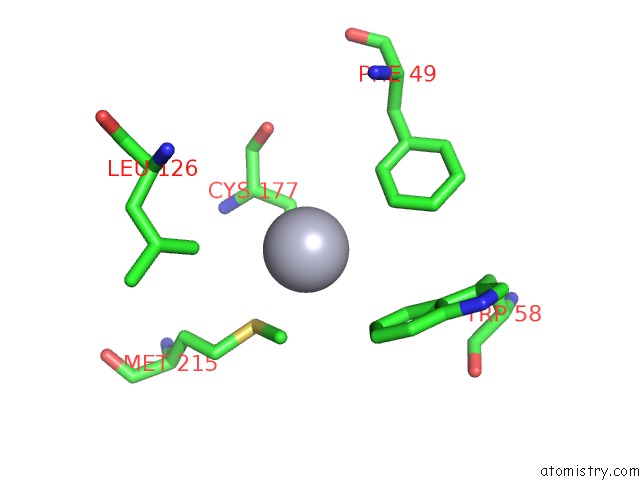
Mercury binding site 3 out of 4 in 1gze
Go back to
Mercury binding site 3 out
of 4 in the Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant)
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 3 of Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant) within 5.0Å range:
|
Mercury binding site 4 out of 4 in 1gze
Go back to
Mercury binding site 4 out
of 4 in the Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant)

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 4 of Structure of the Clostridium Botulinum C3 Exoenzyme (L177C Mutant) within 5.0Å range:
|
Reference:
J.Menetrey,
G.Flatau,
E.A.Stura,
J.B.Charbonnier,
F.Gas,
J.M.Teulon,
M.H.Le Du,
P.Boquet,
A.Menez.
Nad Binding Induces Conformational Changes in Rho Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme J.Biol.Chem. V. 277 30950 2002.
ISSN: ISSN 0021-9258
PubMed: 12029083
DOI: 10.1074/JBC.M201844200
Page generated: Sat Aug 10 23:53:39 2024
ISSN: ISSN 0021-9258
PubMed: 12029083
DOI: 10.1074/JBC.M201844200
Last articles
Cl in 3O4DCl in 3O4B
Cl in 3O3P
Cl in 3O2R
Cl in 3O40
Cl in 3O3Y
Cl in 3O32
Cl in 3O24
Cl in 3O3L
Cl in 3O2K