Mercury »
PDB 1is9-1obh »
1jqc »
Mercury in PDB 1jqc: Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
Enzymatic activity of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
All present enzymatic activity of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine:
1.17.4.1;
1.17.4.1;
Protein crystallography data
The structure of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine, PDB code: 1jqc
was solved by
M.Hogbom,
M.E.Andersson,
P.Nordlund,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.00 / 1.61 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.848, 84.554, 114.286, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.6 / 21.9 |
Other elements in 1jqc:
The structure of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine also contains other interesting chemical elements:
| Manganese | (Mn) | 4 atoms |
Mercury Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 13;Binding sites:
The binding sites of Mercury atom in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine (pdb code 1jqc). This binding sites where shown within 5.0 Angstroms radius around Mercury atom.In total 13 binding sites of Mercury where determined in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine, PDB code: 1jqc:
Jump to Mercury binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Mercury binding site 1 out of 13 in 1jqc
Go back to
Mercury binding site 1 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
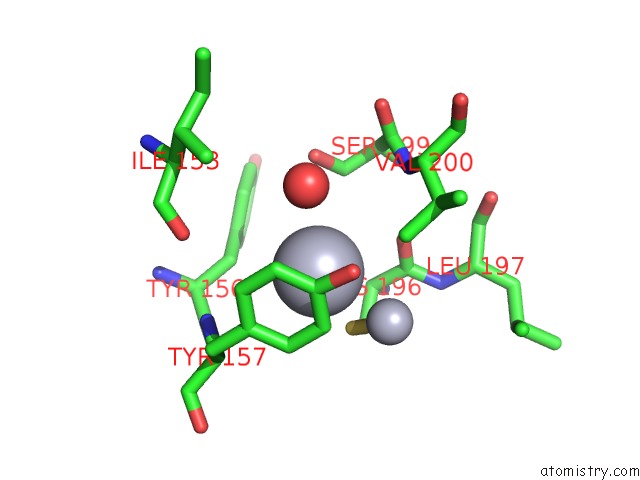
Mono view
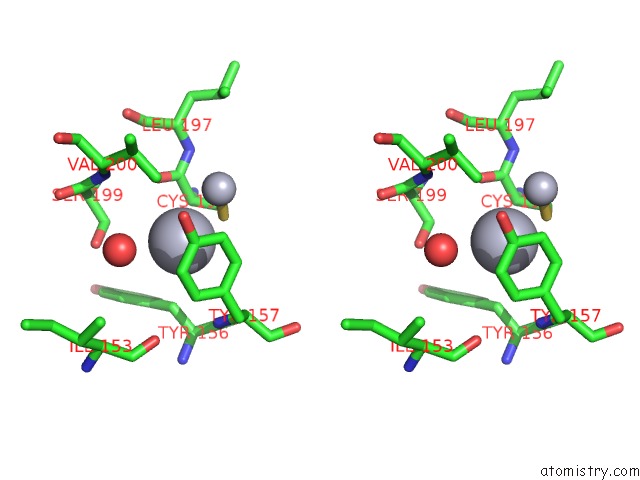
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 2 out of 13 in 1jqc
Go back to
Mercury binding site 2 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
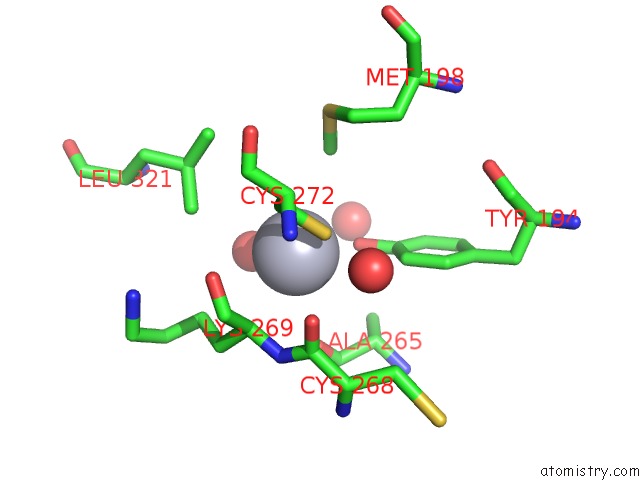
Mono view
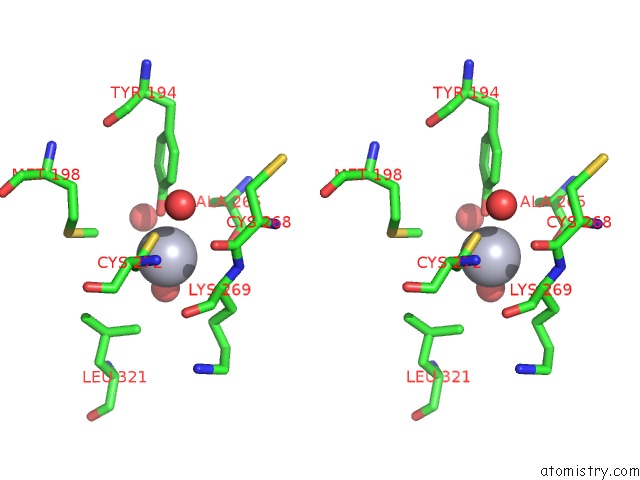
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 3 out of 13 in 1jqc
Go back to
Mercury binding site 3 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
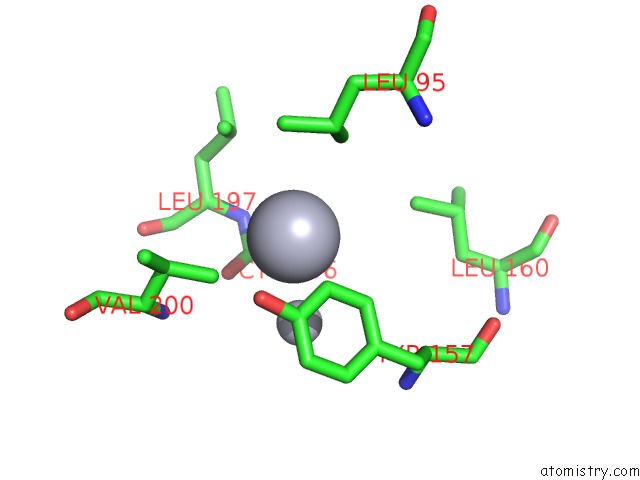
Mono view
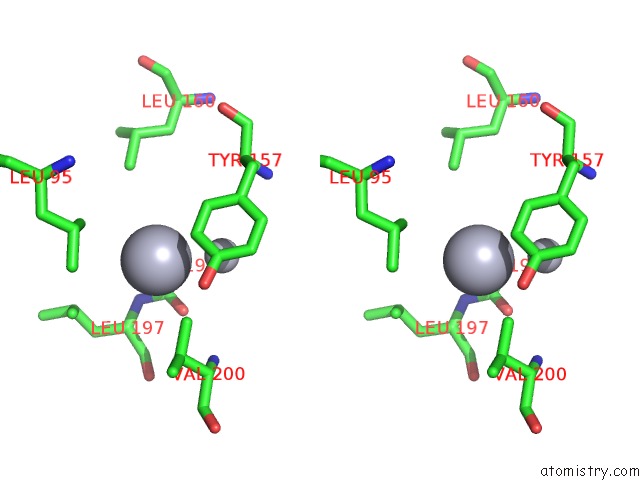
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 3 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 4 out of 13 in 1jqc
Go back to
Mercury binding site 4 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
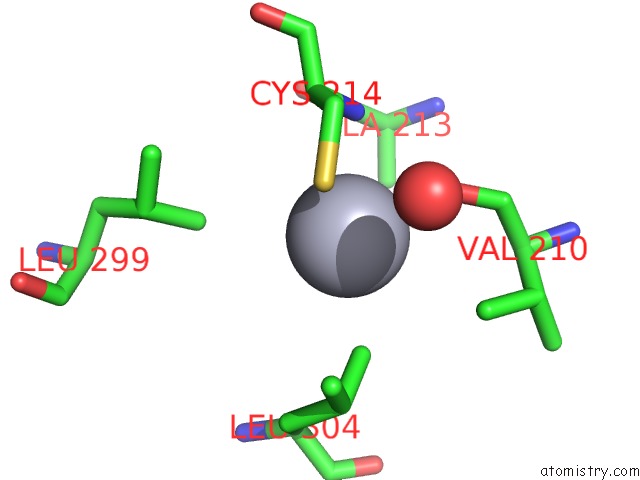
Mono view
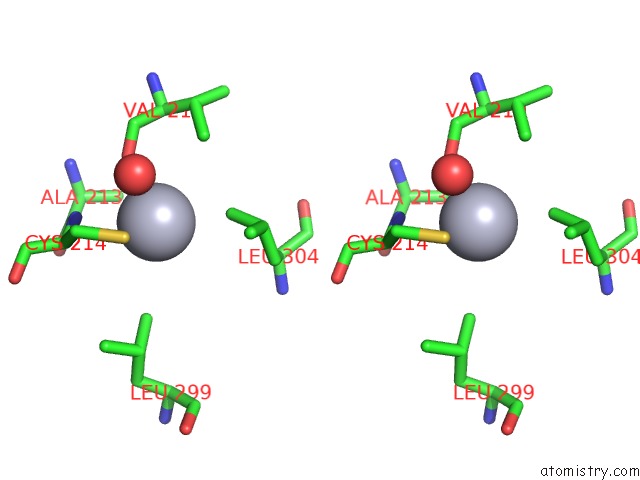
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 4 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 5 out of 13 in 1jqc
Go back to
Mercury binding site 5 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
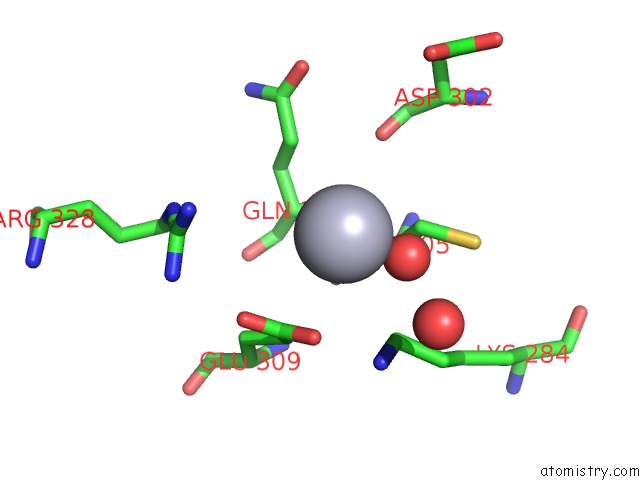
Mono view
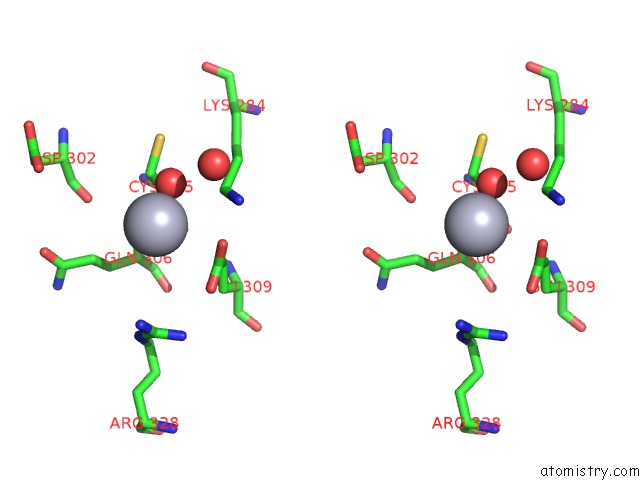
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 5 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 6 out of 13 in 1jqc
Go back to
Mercury binding site 6 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
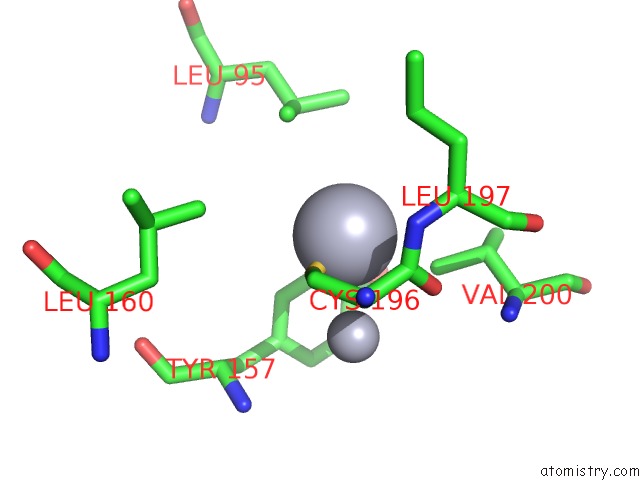
Mono view
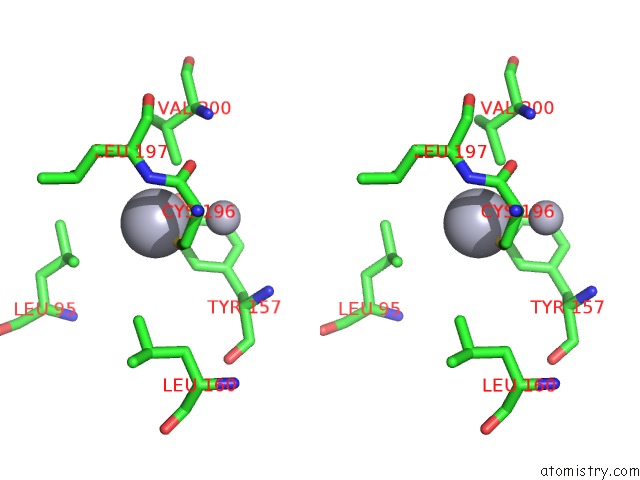
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 6 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 7 out of 13 in 1jqc
Go back to
Mercury binding site 7 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
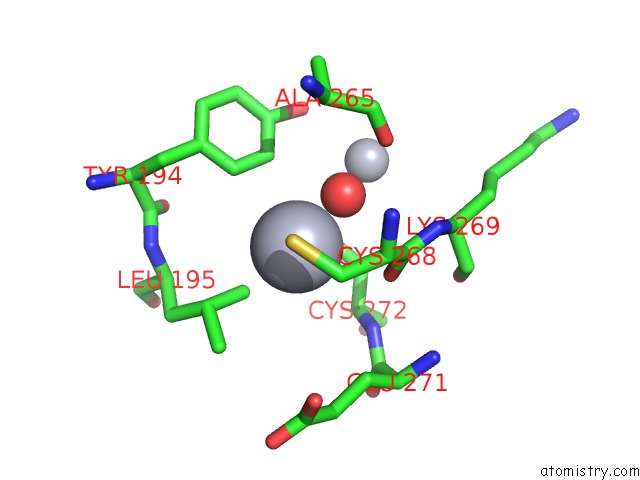
Mono view
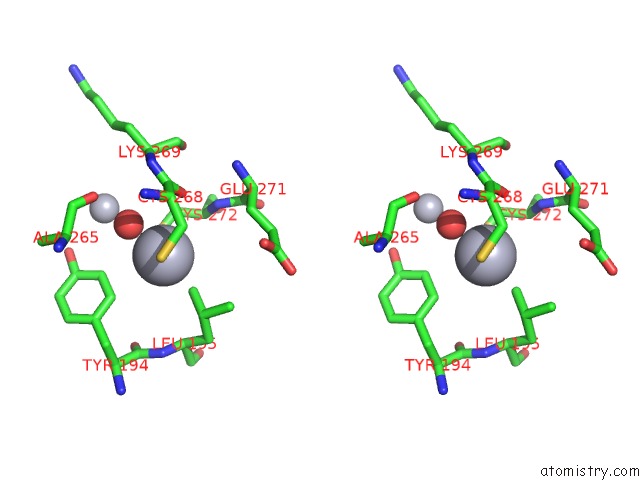
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 7 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 8 out of 13 in 1jqc
Go back to
Mercury binding site 8 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
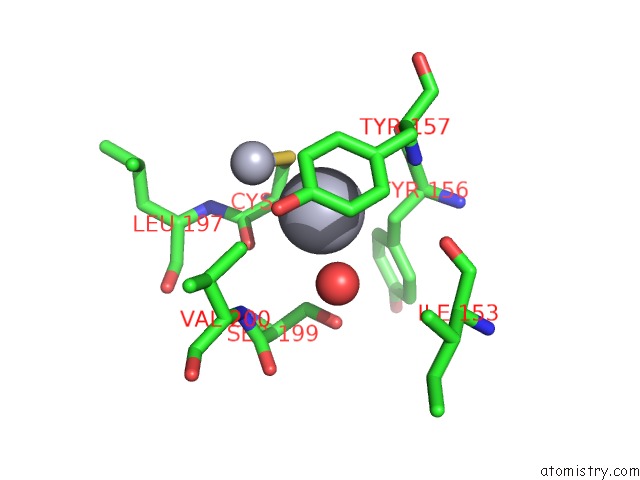
Mono view
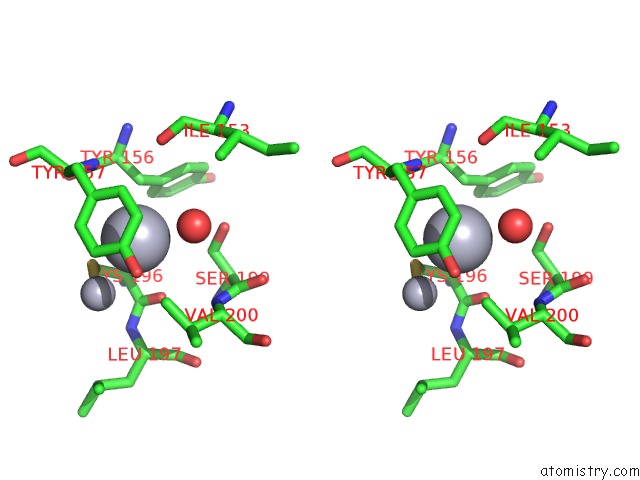
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 8 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 9 out of 13 in 1jqc
Go back to
Mercury binding site 9 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
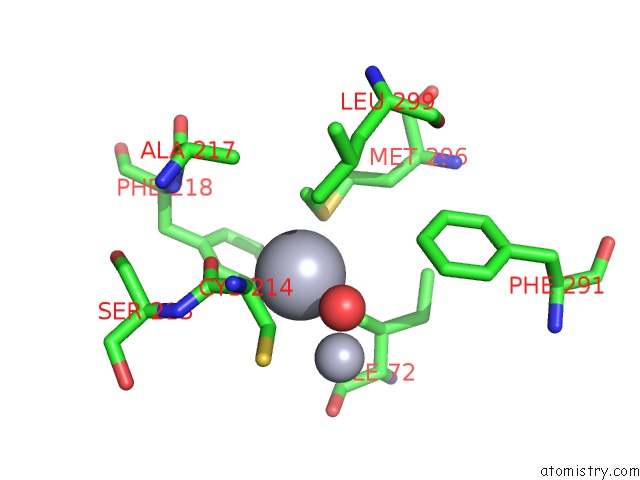
Mono view
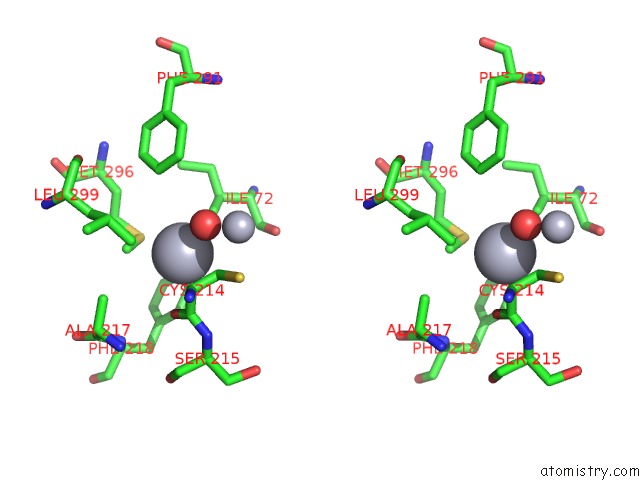
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 9 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Mercury binding site 10 out of 13 in 1jqc
Go back to
Mercury binding site 10 out
of 13 in the Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine
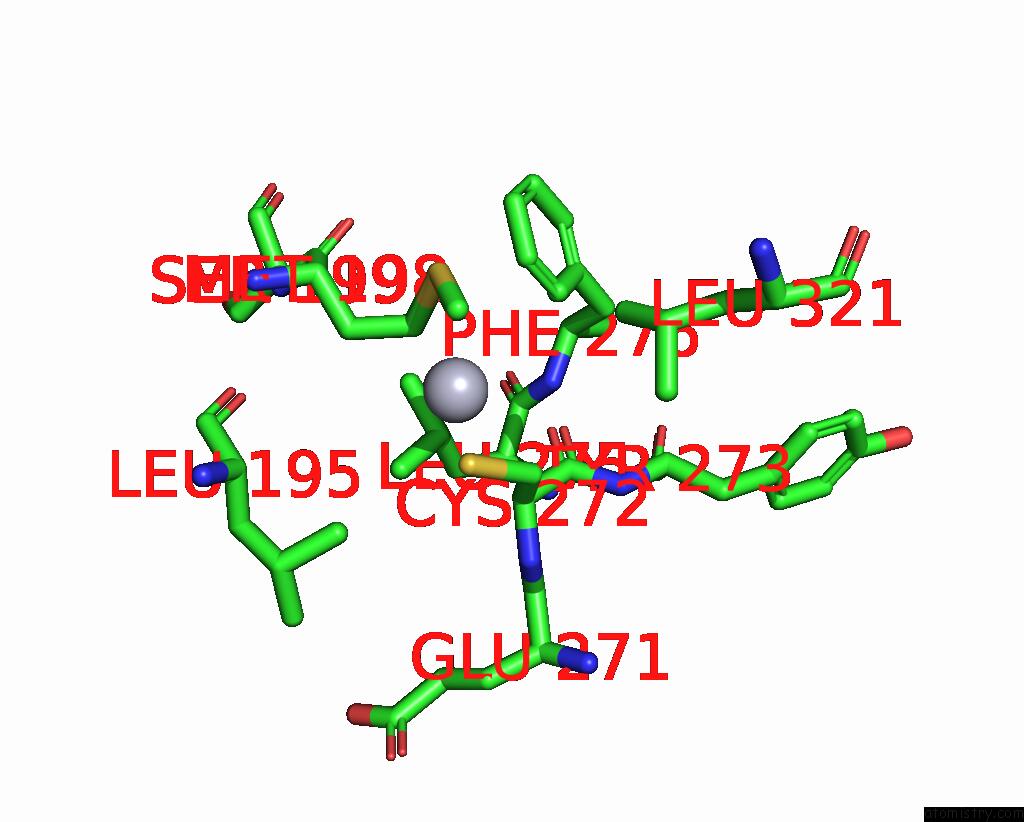
Mono view
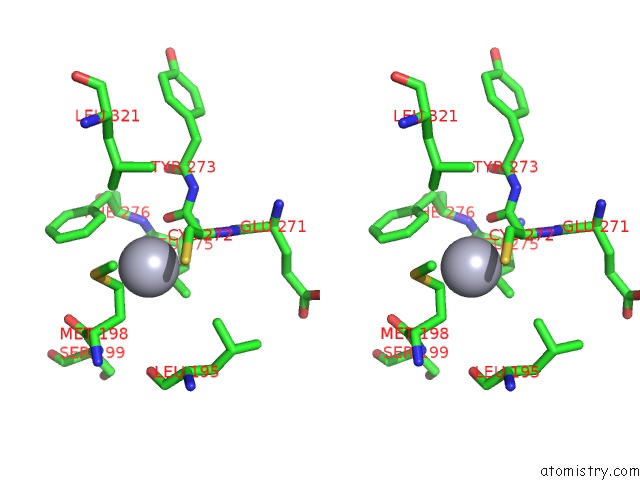
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 10 of Mn Substituted Ribonucleotide Reductase R2 From E. Coli Oxidized By Hydrogen Peroxide and Hydroxylamine within 5.0Å range:
|
Reference:
M.Hogbom,
M.E.Andersson,
P.Nordlund.
Crystal Structures of Oxidized Dinuclear Manganese Centres in Mn-Substituted Class I Ribonucleotide Reductase From Escherichia Coli: Carboxylate Shifts with Implications For O2 Activation and Radical Generation. J.Biol.Inorg.Chem. V. 6 315 2001.
ISSN: ISSN 0949-8257
PubMed: 11315567
DOI: 10.1007/S007750000205
Page generated: Sun Aug 11 00:17:41 2024
ISSN: ISSN 0949-8257
PubMed: 11315567
DOI: 10.1007/S007750000205
Last articles
Cl in 2QO3Cl in 2QOA
Cl in 2QNN
Cl in 2QML
Cl in 2QNL
Cl in 2QMO
Cl in 2QN3
Cl in 2QKF
Cl in 2QLT
Cl in 2QIO