Mercury »
PDB 1is9-1obh »
1obh »
Mercury in PDB 1obh: Leucyl-Trna Synthetase From Thermus Thermophilus Complexed with A Pre-Transfer Editing Substrate Analogue in Both Synthetic Active Site and Editing Site
Protein crystallography data
The structure of Leucyl-Trna Synthetase From Thermus Thermophilus Complexed with A Pre-Transfer Editing Substrate Analogue in Both Synthetic Active Site and Editing Site, PDB code: 1obh
was solved by
S.Cusack,
A.Yaremchuk,
M.Tukalo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.99 / 2.2 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 103.515, 152.884, 172.605, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.3 / 23.4 |
Mercury Binding Sites:
The binding sites of Mercury atom in the Leucyl-Trna Synthetase From Thermus Thermophilus Complexed with A Pre-Transfer Editing Substrate Analogue in Both Synthetic Active Site and Editing Site
(pdb code 1obh). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total only one binding site of Mercury was determined in the Leucyl-Trna Synthetase From Thermus Thermophilus Complexed with A Pre-Transfer Editing Substrate Analogue in Both Synthetic Active Site and Editing Site, PDB code: 1obh:
In total only one binding site of Mercury was determined in the Leucyl-Trna Synthetase From Thermus Thermophilus Complexed with A Pre-Transfer Editing Substrate Analogue in Both Synthetic Active Site and Editing Site, PDB code: 1obh:
Mercury binding site 1 out of 1 in 1obh
Go back to
Mercury binding site 1 out
of 1 in the Leucyl-Trna Synthetase From Thermus Thermophilus Complexed with A Pre-Transfer Editing Substrate Analogue in Both Synthetic Active Site and Editing Site
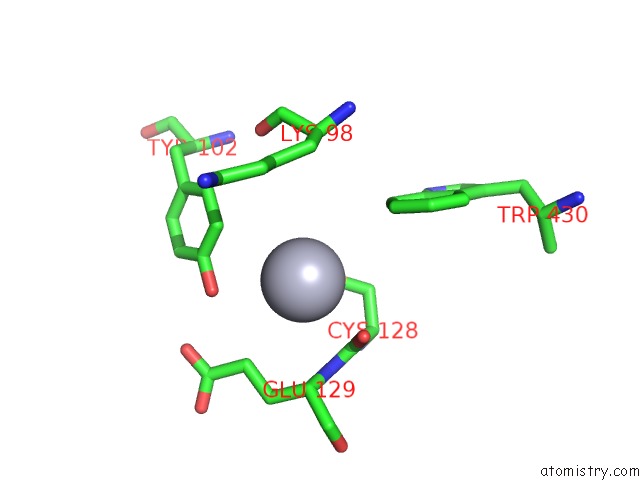
Mono view
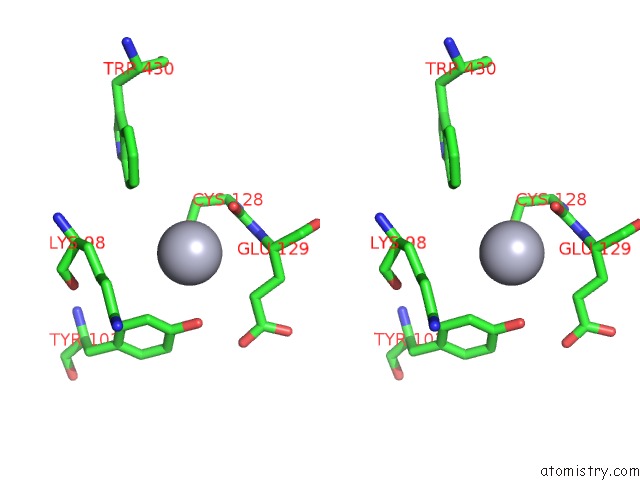
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Leucyl-Trna Synthetase From Thermus Thermophilus Complexed with A Pre-Transfer Editing Substrate Analogue in Both Synthetic Active Site and Editing Site within 5.0Å range:
|
Reference:
T.Lincecum,
M.Tukalo,
A.Yaremchuk,
R.Mursinna,
A.Williams,
B.Sproat,
W.Van Den Eynde,
A.Link,
S.Van Calenbergh,
M.Grotli,
S.Martinis,
S.Cusack.
Structural and Mechanistic Basis of Pre- and Posttransfer Editing By Leucyl-Trna Synthetase Mol.Cell V. 11 951 2003.
ISSN: ISSN 1097-2765
PubMed: 12718881
DOI: 10.1016/S1097-2765(03)00098-4
Page generated: Sun Aug 11 00:45:55 2024
ISSN: ISSN 1097-2765
PubMed: 12718881
DOI: 10.1016/S1097-2765(03)00098-4
Last articles
F in 4F2AF in 4F2Y
F in 4E99
F in 4F2X
F in 4EZJ
F in 4EWS
F in 4ELF
F in 4EWQ
F in 4EQU
F in 4EST