Mercury »
PDB 1of5-1rsr »
1r82 »
Mercury in PDB 1r82: Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose
Enzymatic activity of Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose
All present enzymatic activity of Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose:
2.4.1.37;
2.4.1.37;
Protein crystallography data
The structure of Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose, PDB code: 1r82
was solved by
H.P.Nguyen,
N.O.L.Seto,
Y.Cai,
E.K.Leinala,
S.N.Borisova,
M.M.Palcic,
S.V.Evans,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.82 / 1.55 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.800, 150.400, 79.500, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.2 / 25.8 |
Mercury Binding Sites:
The binding sites of Mercury atom in the Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose
(pdb code 1r82). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 2 binding sites of Mercury where determined in the Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose, PDB code: 1r82:
Jump to Mercury binding site number: 1; 2;
In total 2 binding sites of Mercury where determined in the Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose, PDB code: 1r82:
Jump to Mercury binding site number: 1; 2;
Mercury binding site 1 out of 2 in 1r82
Go back to
Mercury binding site 1 out
of 2 in the Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose
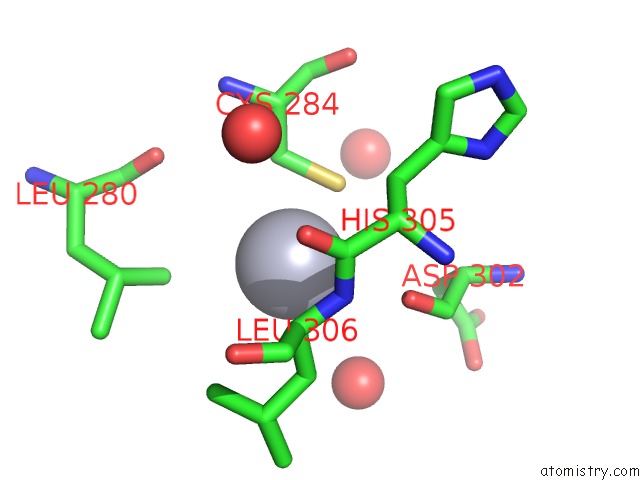
Mono view
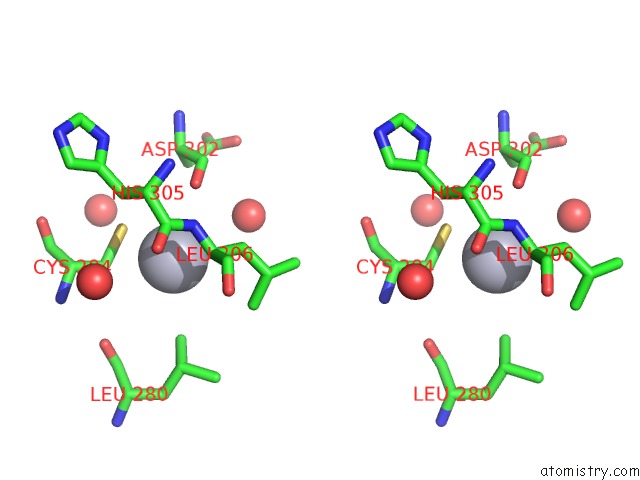
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose within 5.0Å range:
|
Mercury binding site 2 out of 2 in 1r82
Go back to
Mercury binding site 2 out
of 2 in the Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose
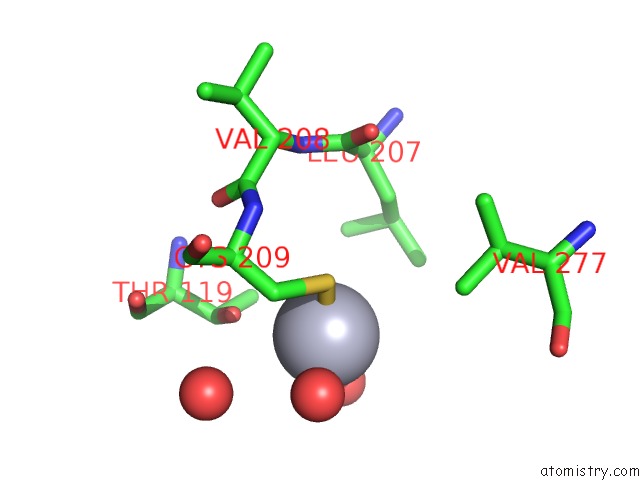
Mono view
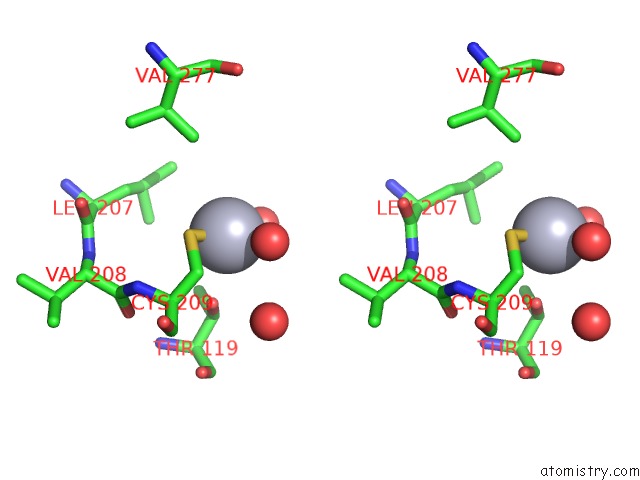
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Glycosyltransferase B in Complex with 3-Amino-Acceptor Analog Inhibitor, and Uridine Diphosphate-Galactose within 5.0Å range:
|
Reference:
H.P.Nguyen,
N.O.L.Seto,
Y.Cai,
E.K.Leinala,
S.N.Borisova,
M.M.Palcic,
S.V.Evans.
The Influence of An Intramolecular Hydrogen Bond in Differential Recognition of Inhibitory Acceptor Analogs By Human Abo(H) Blood Group A and B Glycosyltransferases J.Biol.Chem. V. 278 49191 2003.
ISSN: ISSN 0021-9258
PubMed: 12972418
DOI: 10.1074/JBC.M308770200
Page generated: Fri Aug 8 09:30:28 2025
ISSN: ISSN 0021-9258
PubMed: 12972418
DOI: 10.1074/JBC.M308770200
Last articles
I in 3S1SI in 3S99
I in 3SD0
I in 3RU6
I in 3S43
I in 3S5Q
I in 3S53
I in 3S2O
I in 3RVI
I in 3S2H