Mercury »
PDB 1rsv-1yp2 »
1s1f »
Mercury in PDB 1s1f: Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways
Protein crystallography data
The structure of Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways, PDB code: 1s1f
was solved by
B.Zhao,
D.C.Lamb,
L.Lei,
M.Sundaramoorthy,
L.M.Podust,
M.R.Waterman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.26 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.250, 65.553, 104.245, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.2 / 21.6 |
Other elements in 1s1f:
The structure of Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways also contains other interesting chemical elements:
| Iron | (Fe) | 1 atom |
Mercury Binding Sites:
The binding sites of Mercury atom in the Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways
(pdb code 1s1f). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 2 binding sites of Mercury where determined in the Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways, PDB code: 1s1f:
Jump to Mercury binding site number: 1; 2;
In total 2 binding sites of Mercury where determined in the Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways, PDB code: 1s1f:
Jump to Mercury binding site number: 1; 2;
Mercury binding site 1 out of 2 in 1s1f
Go back to
Mercury binding site 1 out
of 2 in the Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways
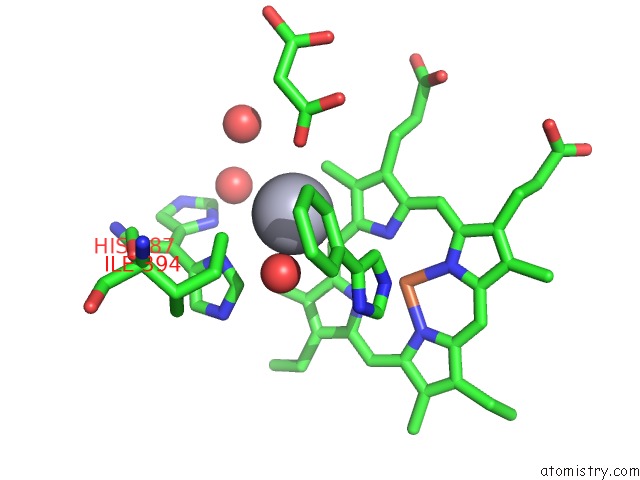
Mono view
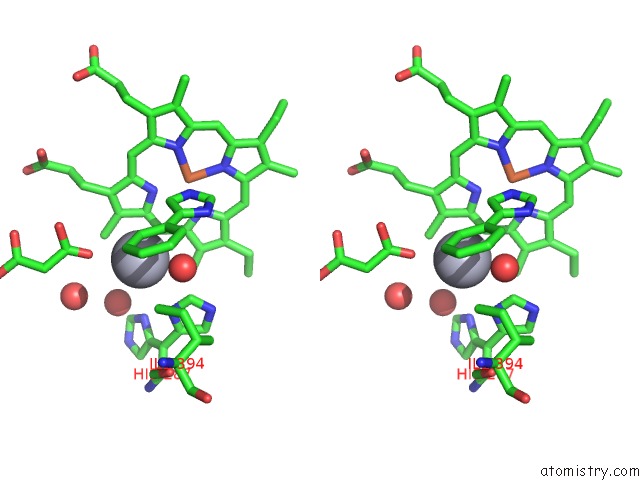
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways within 5.0Å range:
|
Mercury binding site 2 out of 2 in 1s1f
Go back to
Mercury binding site 2 out
of 2 in the Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways
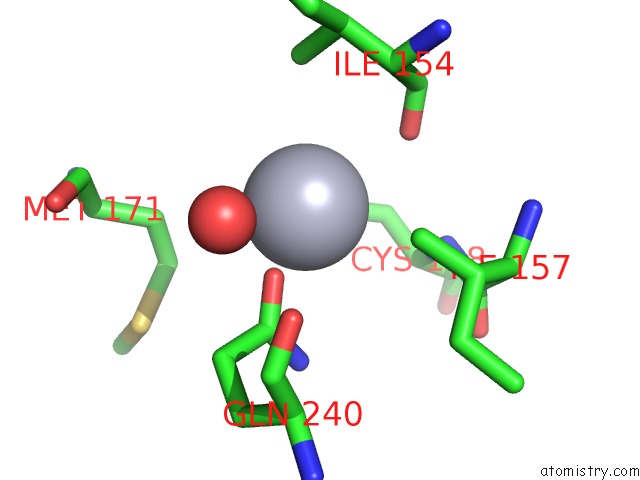
Mono view
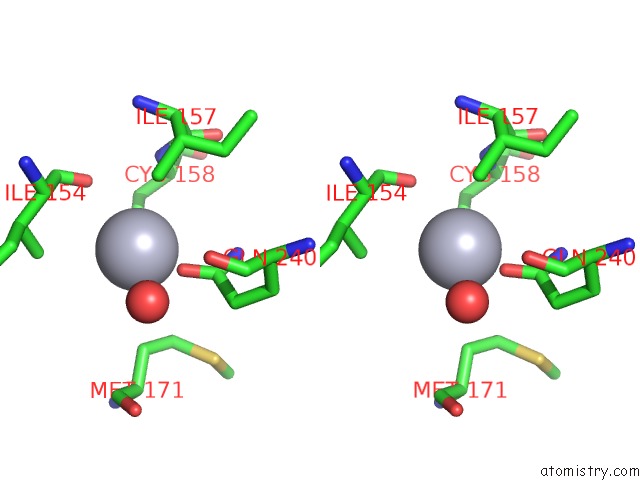
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 From Antibiotic Biosynthetic Pathways within 5.0Å range:
|
Reference:
B.Zhao,
F.P.Guengerich,
A.Bellamine,
D.C.Lamb,
M.Izumikawa,
L.Lei,
L.M.Podust,
M.Sundaramoorthy,
J.A.Kalaitzis,
L.M.Reddy,
S.L.Kelly,
B.S.Moore,
D.Stec,
M.Voehler,
J.R.Falck,
T.Shimada,
M.R.Waterman.
Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces Coelicolor A3(2) Cytochrome P450 158A2 J.Biol.Chem. V. 280 11599 2005.
ISSN: ISSN 0021-9258
PubMed: 15659395
DOI: 10.1074/JBC.M410933200
Page generated: Sun Aug 11 01:39:34 2024
ISSN: ISSN 0021-9258
PubMed: 15659395
DOI: 10.1074/JBC.M410933200
Last articles
Ca in 5MFGCa in 5MFA
Ca in 5MF4
Ca in 5MEY
Ca in 5MB1
Ca in 5MAZ
Ca in 5MER
Ca in 5MAY
Ca in 5MEH
Ca in 5MC9