Mercury »
PDB 1rsv-1yp2 »
1x8k »
Mercury in PDB 1x8k: Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap
Protein crystallography data
The structure of Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap, PDB code: 1x8k
was solved by
S.Pakhomova,
J.Buck,
M.E.Newcomer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 15.00 / 2.75 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.286, 66.661, 84.464, 90.00, 110.66, 90.00 |
| R / Rfree (%) | 19.4 / 25.5 |
Other elements in 1x8k:
The structure of Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
Mercury Binding Sites:
The binding sites of Mercury atom in the Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap
(pdb code 1x8k). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 2 binding sites of Mercury where determined in the Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap, PDB code: 1x8k:
Jump to Mercury binding site number: 1; 2;
In total 2 binding sites of Mercury where determined in the Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap, PDB code: 1x8k:
Jump to Mercury binding site number: 1; 2;
Mercury binding site 1 out of 2 in 1x8k
Go back to
Mercury binding site 1 out
of 2 in the Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap
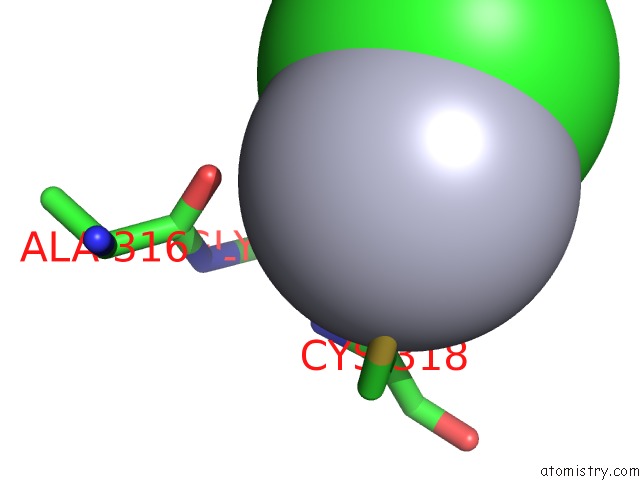
Mono view
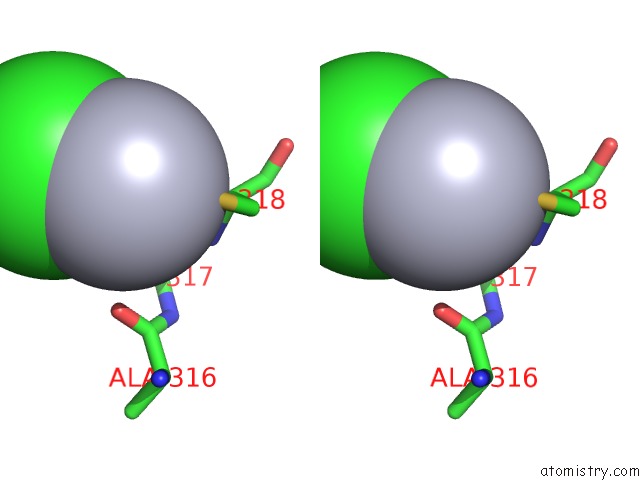
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap within 5.0Å range:
|
Mercury binding site 2 out of 2 in 1x8k
Go back to
Mercury binding site 2 out
of 2 in the Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap
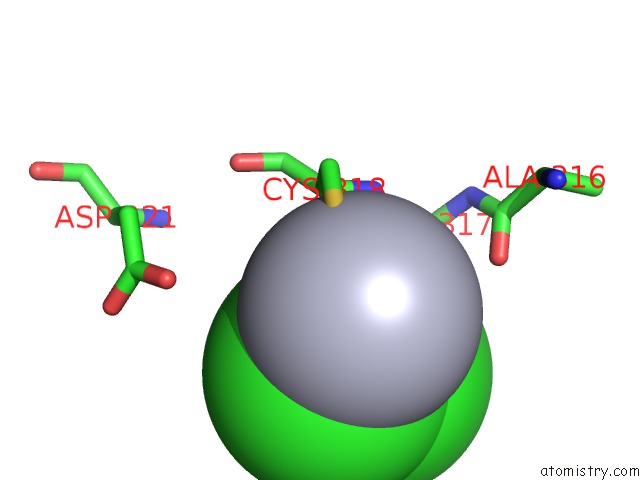
Mono view
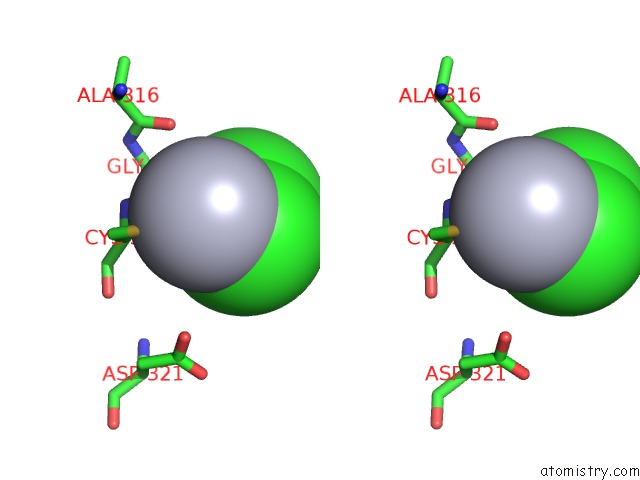
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Crystal Structure of Retinol Dehydratase in Complex with Anhydroretinol and Inactive Cofactor Pap within 5.0Å range:
|
Reference:
S.Pakhomova,
J.Buck,
M.E.Newcomer.
The Structures of the Unique Sulfotransferase Retinol Dehydratase with Product and Inhibitors Provide Insight Into Enzyme Mechanism and Inhibition. Protein Sci. V. 14 176 2005.
ISSN: ISSN 0961-8368
PubMed: 15608121
DOI: 10.1110/PS.041061105
Page generated: Sun Aug 11 01:51:52 2024
ISSN: ISSN 0961-8368
PubMed: 15608121
DOI: 10.1110/PS.041061105
Last articles
Cl in 7UUJCl in 7UUG
Cl in 7USZ
Cl in 7UP4
Cl in 7UOS
Cl in 7UPI
Cl in 7UMO
Cl in 7UOD
Cl in 7UN0
Cl in 7UOQ