Mercury »
PDB 2esw-2o1f »
2ga8 »
Mercury in PDB 2ga8: Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation.
Protein crystallography data
The structure of Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation., PDB code: 2ga8
was solved by
V.Chaptal,
S.Morera,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.77 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 135.278, 135.278, 48.476, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.4 / 21.9 |
Mercury Binding Sites:
The binding sites of Mercury atom in the Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation.
(pdb code 2ga8). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 4 binding sites of Mercury where determined in the Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation., PDB code: 2ga8:
Jump to Mercury binding site number: 1; 2; 3; 4;
In total 4 binding sites of Mercury where determined in the Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation., PDB code: 2ga8:
Jump to Mercury binding site number: 1; 2; 3; 4;
Mercury binding site 1 out of 4 in 2ga8
Go back to
Mercury binding site 1 out
of 4 in the Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation.
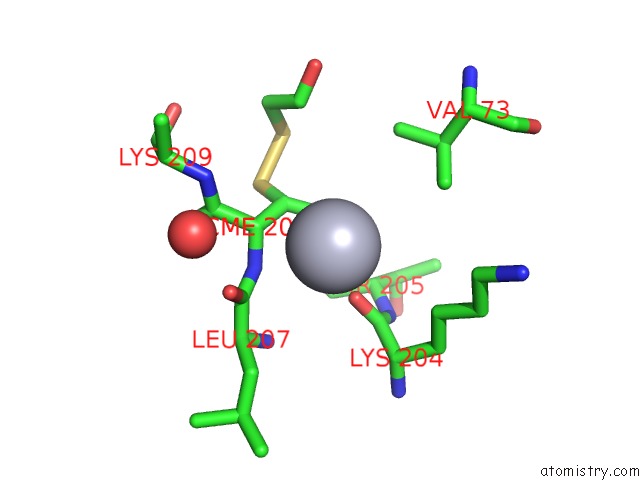
Mono view
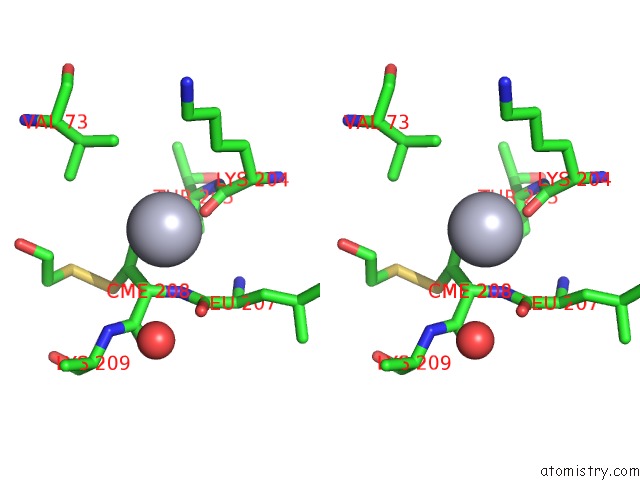
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation. within 5.0Å range:
|
Mercury binding site 2 out of 4 in 2ga8
Go back to
Mercury binding site 2 out
of 4 in the Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation.
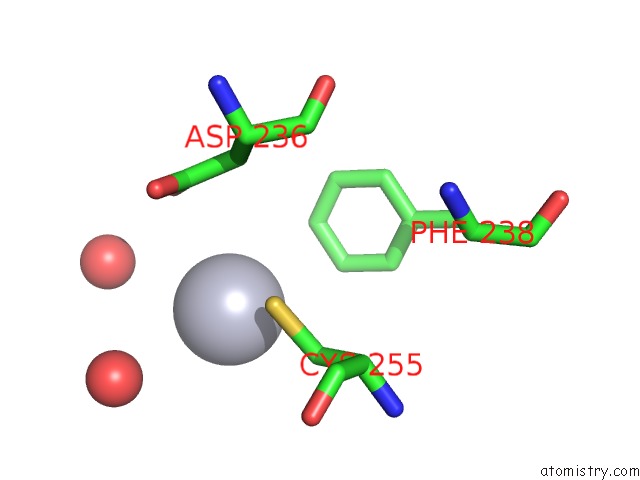
Mono view
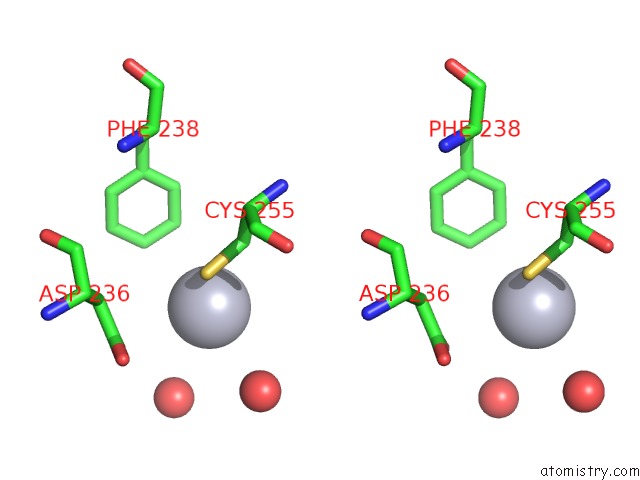
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation. within 5.0Å range:
|
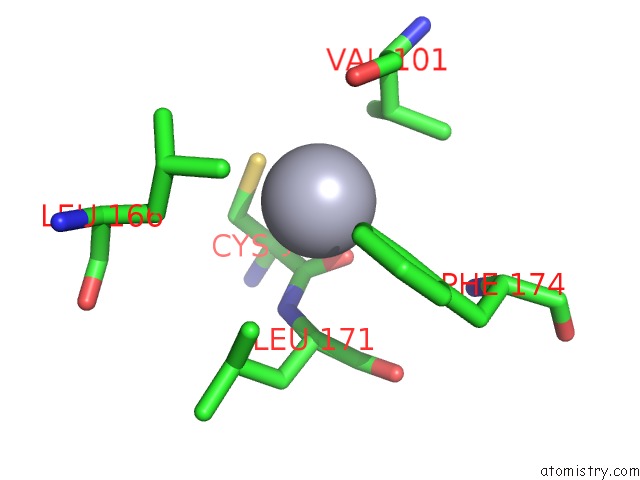
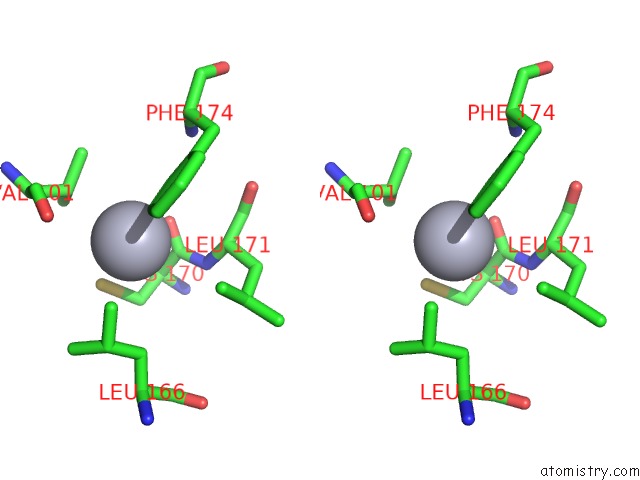
Mercury binding site 3 out of 4 in 2ga8
Go back to
Mercury binding site 3 out
of 4 in the Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 3 of Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation. within 5.0Å range:
|
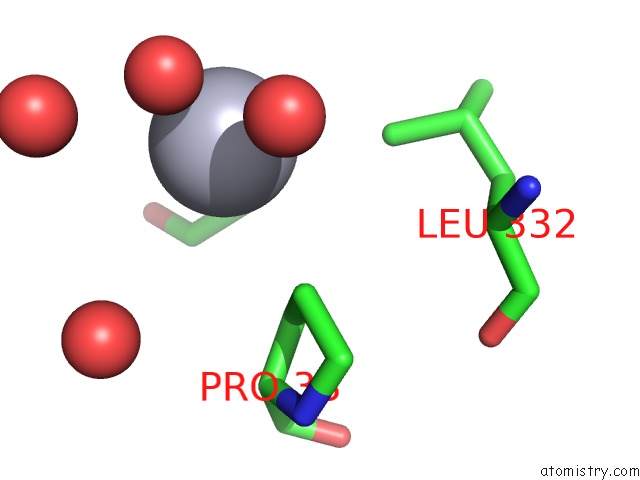
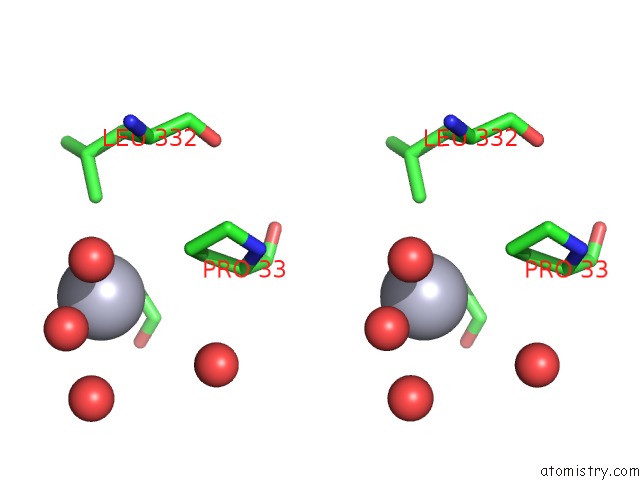
Mercury binding site 4 out of 4 in 2ga8
Go back to
Mercury binding site 4 out
of 4 in the Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 4 of Crystal Structure of YFH7 From Saccharomyces Cerevisiae: A Putative P-Loop Containing Kinase with A Circular Permutation. within 5.0Å range:
|
Reference:
V.Gueguen-Chaignon,
V.Chaptal,
L.Lariviere,
N.Costa,
P.Lopes,
S.Morera,
S.Nessler.
Crystal Structure and Functional Analysis Identify the P-Loop Containing Protein YFH7 of Saccharomyces Cerevisiae As An Atp-Dependent Kinase. Proteins V. 71 804 2007.
ISSN: ISSN 0887-3585
PubMed: 18004758
DOI: 10.1002/PROT.21740
Page generated: Sun Aug 11 02:35:01 2024
ISSN: ISSN 0887-3585
PubMed: 18004758
DOI: 10.1002/PROT.21740
Last articles
F in 3WGVF in 3WV1
F in 3WGU
F in 3WRU
F in 3WPN
F in 3WK8
F in 3WIG
F in 3W9S
F in 3WFG
F in 3WFF