Mercury »
PDB 2o1g-3b4f »
2w9m »
Mercury in PDB 2w9m: Structure of Family X Dna Polymerase From Deinococcus Radiodurans
Protein crystallography data
The structure of Structure of Family X Dna Polymerase From Deinococcus Radiodurans, PDB code: 2w9m
was solved by
N.Leulliot,
L.Cladiere,
F.Lecointe,
D.Durand,
U.Hubscher,
H.Van Tilbeurgh,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.32 / 2.46 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.930, 138.250, 67.750, 90.00, 92.25, 90.00 |
| R / Rfree (%) | 20.3 / 24.9 |
Other elements in 2w9m:
The structure of Structure of Family X Dna Polymerase From Deinococcus Radiodurans also contains other interesting chemical elements:
| Zinc | (Zn) | 7 atoms |
Mercury Binding Sites:
The binding sites of Mercury atom in the Structure of Family X Dna Polymerase From Deinococcus Radiodurans
(pdb code 2w9m). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 2 binding sites of Mercury where determined in the Structure of Family X Dna Polymerase From Deinococcus Radiodurans, PDB code: 2w9m:
Jump to Mercury binding site number: 1; 2;
In total 2 binding sites of Mercury where determined in the Structure of Family X Dna Polymerase From Deinococcus Radiodurans, PDB code: 2w9m:
Jump to Mercury binding site number: 1; 2;
Mercury binding site 1 out of 2 in 2w9m
Go back to
Mercury binding site 1 out
of 2 in the Structure of Family X Dna Polymerase From Deinococcus Radiodurans
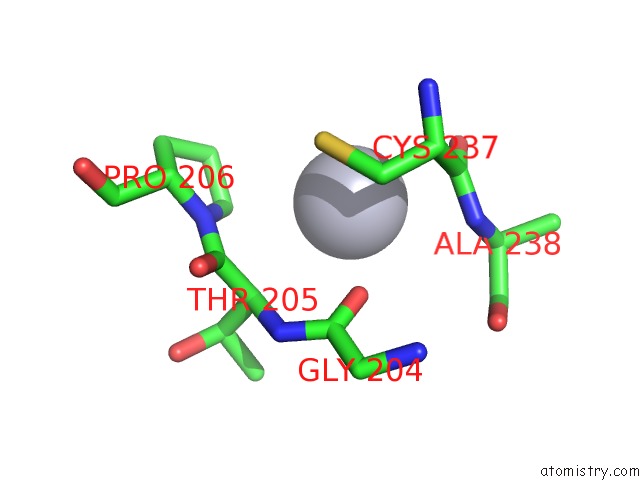
Mono view
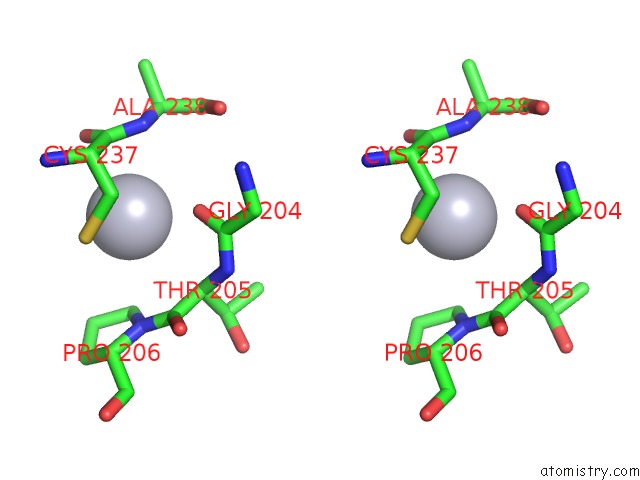
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Structure of Family X Dna Polymerase From Deinococcus Radiodurans within 5.0Å range:
|
Mercury binding site 2 out of 2 in 2w9m
Go back to
Mercury binding site 2 out
of 2 in the Structure of Family X Dna Polymerase From Deinococcus Radiodurans
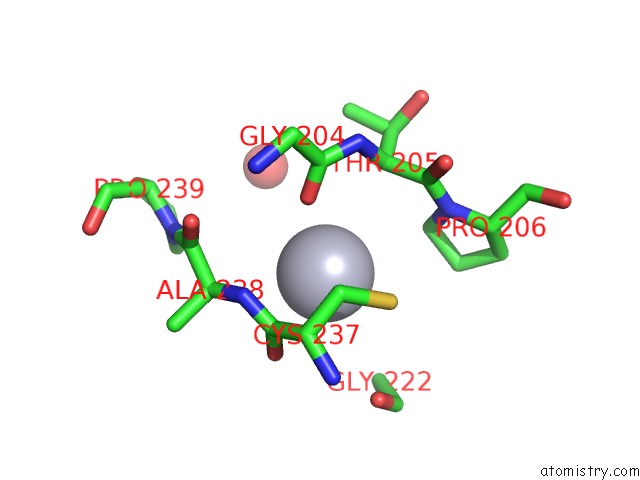
Mono view
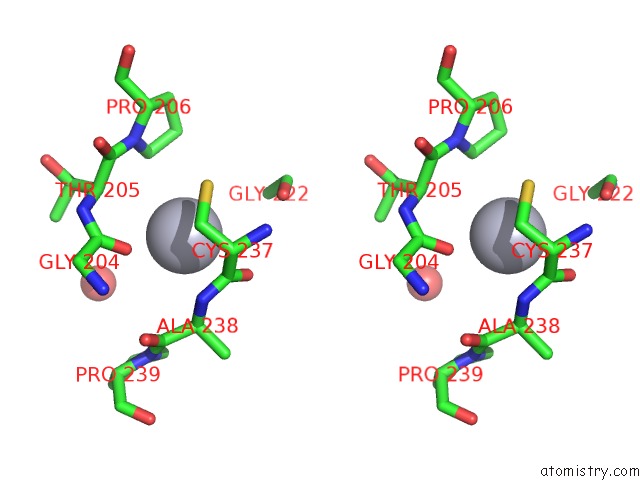
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Structure of Family X Dna Polymerase From Deinococcus Radiodurans within 5.0Å range:
|
Reference:
N.Leulliot,
L.Cladiere,
F.Lecointe,
D.Durand,
U.Hubscher,
H.Van Tilbeurgh.
The Family X Dna Polymerase From Deinococcus Radioduran Adopts A Non-Standard Extended Conformation. J.Biol.Chem. V. 284 11992 2009.
ISSN: ISSN 0021-9258
PubMed: 19251692
DOI: 10.1074/JBC.M809342200
Page generated: Sun Aug 11 03:02:43 2024
ISSN: ISSN 0021-9258
PubMed: 19251692
DOI: 10.1074/JBC.M809342200
Last articles
F in 7MYUF in 7MYR
F in 7MYO
F in 7MXN
F in 7MXG
F in 7MXH
F in 7MX7
F in 7MVS
F in 7MX0
F in 7MXA