Mercury »
PDB 3kbc-3wa8 »
3vus »
Mercury in PDB 3vus: Escherichia Coli Pgab N-Terminal Domain
Protein crystallography data
The structure of Escherichia Coli Pgab N-Terminal Domain, PDB code: 3vus
was solved by
T.Nishiyama,
H.Noguchi,
H.Yoshida,
S.-Y.Park,
J.R.H.Tame,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 1.65 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.559, 53.114, 144.170, 90.00, 95.25, 90.00 |
| R / Rfree (%) | 20.5 / 25.6 |
Other elements in 3vus:
The structure of Escherichia Coli Pgab N-Terminal Domain also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Mercury Binding Sites:
The binding sites of Mercury atom in the Escherichia Coli Pgab N-Terminal Domain
(pdb code 3vus). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total only one binding site of Mercury was determined in the Escherichia Coli Pgab N-Terminal Domain, PDB code: 3vus:
In total only one binding site of Mercury was determined in the Escherichia Coli Pgab N-Terminal Domain, PDB code: 3vus:
Mercury binding site 1 out of 1 in 3vus
Go back to
Mercury binding site 1 out
of 1 in the Escherichia Coli Pgab N-Terminal Domain
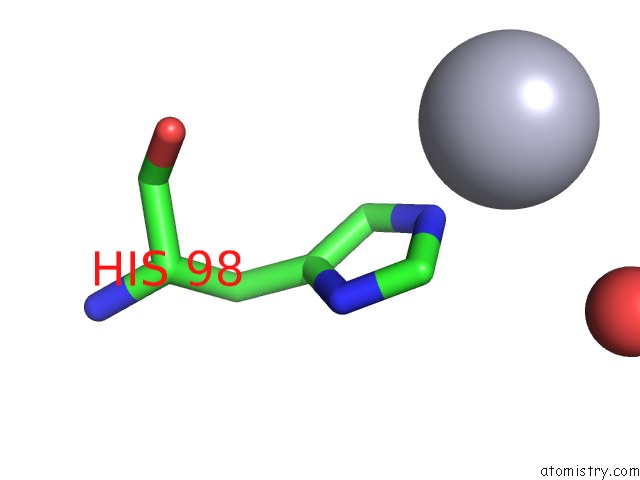
Mono view
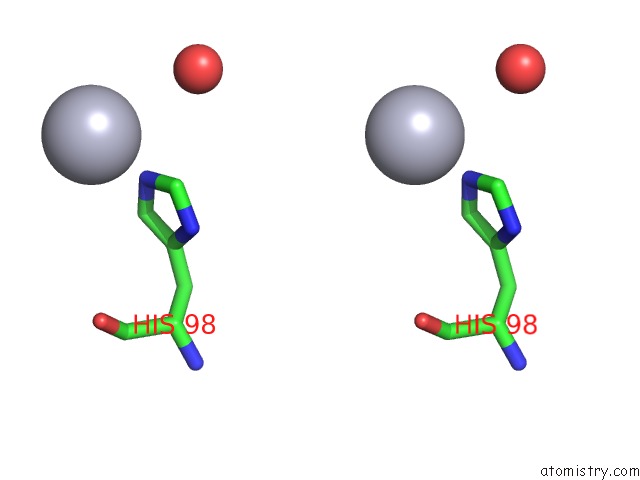
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Escherichia Coli Pgab N-Terminal Domain within 5.0Å range:
|
Reference:
T.Nishiyama,
H.Noguchi,
H.Yoshida,
S.Y.Park,
J.R.Tame.
The Structure of the Deacetylase Domain of Escherichia Coli Pgab, An Enzyme Required For Biofilm Formation: A Circularly Permuted Member of the Carbohydrate Esterase 4 Family Acta Crystallogr.,Sect.D V. 69 44 2013.
ISSN: ISSN 0907-4449
PubMed: 23275162
DOI: 10.1107/S0907444912042059
Page generated: Sun Aug 11 04:11:13 2024
ISSN: ISSN 0907-4449
PubMed: 23275162
DOI: 10.1107/S0907444912042059
Last articles
Cl in 7U0ACl in 7TZX
Cl in 7TZW
Cl in 7TZY
Cl in 7TXR
Cl in 7TZ6
Cl in 7TZN
Cl in 7TZM
Cl in 7TXN
Cl in 7TXO