Mercury »
PDB 5ca2-5l9w »
5fta »
Mercury in PDB 5fta: Crystal Structure of the N-Terminal Btb Domain of Human KCTD10
Protein crystallography data
The structure of Crystal Structure of the N-Terminal Btb Domain of Human KCTD10, PDB code: 5fta
was solved by
D.M.Pinkas,
C.E.Sanvitale,
N.Solcan,
S.Goubin,
C.Tallant,
J.A.Newman,
J.Kopec,
F.Fitzpatrick,
R.Talon,
P.Collins,
T.Krojer,
F.Von Delft,
C.H.Arrowsmith,
A.M.Edwards,
C.Bountra,
A.Bullock,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 17.77 / 2.64 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 48.150, 83.660, 58.680, 90.00, 95.18, 90.00 |
| R / Rfree (%) | 22.6 / 25.8 |
Mercury Binding Sites:
The binding sites of Mercury atom in the Crystal Structure of the N-Terminal Btb Domain of Human KCTD10
(pdb code 5fta). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 4 binding sites of Mercury where determined in the Crystal Structure of the N-Terminal Btb Domain of Human KCTD10, PDB code: 5fta:
Jump to Mercury binding site number: 1; 2; 3; 4;
In total 4 binding sites of Mercury where determined in the Crystal Structure of the N-Terminal Btb Domain of Human KCTD10, PDB code: 5fta:
Jump to Mercury binding site number: 1; 2; 3; 4;
Mercury binding site 1 out of 4 in 5fta
Go back to
Mercury binding site 1 out
of 4 in the Crystal Structure of the N-Terminal Btb Domain of Human KCTD10

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Crystal Structure of the N-Terminal Btb Domain of Human KCTD10 within 5.0Å range:
|
Mercury binding site 2 out of 4 in 5fta
Go back to
Mercury binding site 2 out
of 4 in the Crystal Structure of the N-Terminal Btb Domain of Human KCTD10

Mono view
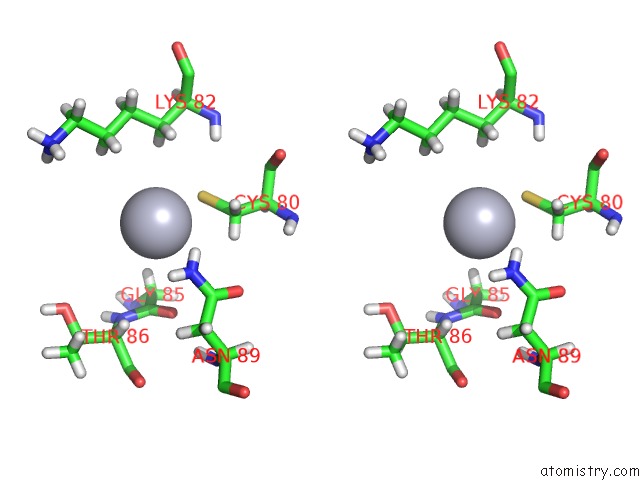
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Crystal Structure of the N-Terminal Btb Domain of Human KCTD10 within 5.0Å range:
|
Mercury binding site 3 out of 4 in 5fta
Go back to
Mercury binding site 3 out
of 4 in the Crystal Structure of the N-Terminal Btb Domain of Human KCTD10
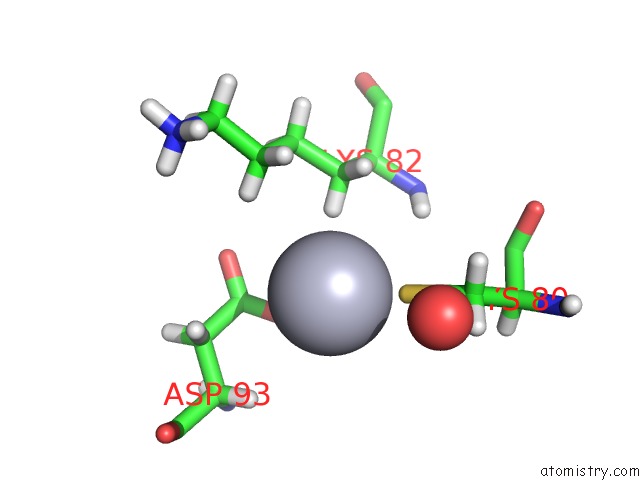
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 3 of Crystal Structure of the N-Terminal Btb Domain of Human KCTD10 within 5.0Å range:
|
Mercury binding site 4 out of 4 in 5fta
Go back to
Mercury binding site 4 out
of 4 in the Crystal Structure of the N-Terminal Btb Domain of Human KCTD10
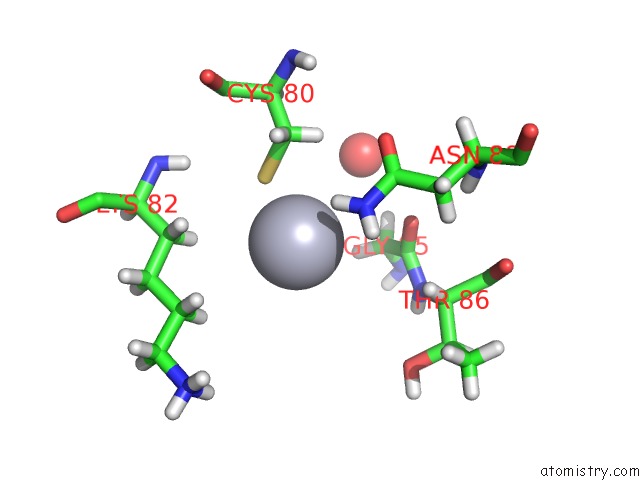
Mono view
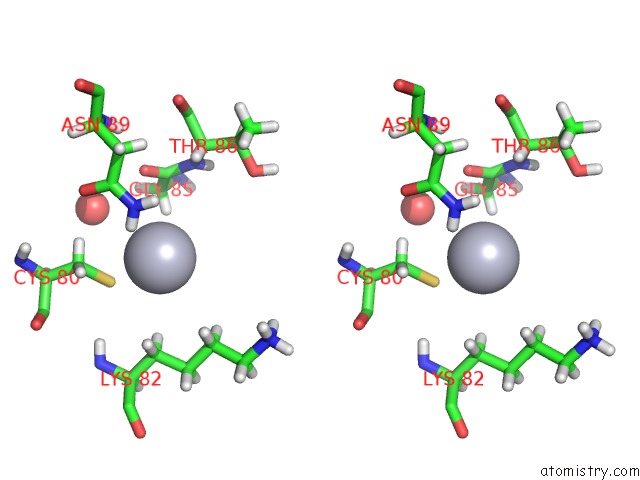
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 4 of Crystal Structure of the N-Terminal Btb Domain of Human KCTD10 within 5.0Å range:
|
Reference:
D.M.Pinkas,
C.E.Sanvitale,
J.C.Bufton,
F.J.Sorrell,
N.Solcan,
R.Chalk,
J.Doutch,
A.N.Bullock.
Structural Complexity in the Kctd Family of CULLIN3-Dependent E3 Ubiquitin Ligases. Biochem. J. V. 474 3747 2017.
ISSN: ESSN 1470-8728
PubMed: 28963344
DOI: 10.1042/BCJ20170527
Page generated: Sun Aug 11 06:21:14 2024
ISSN: ESSN 1470-8728
PubMed: 28963344
DOI: 10.1042/BCJ20170527
Last articles
Ca in 5TEACa in 5TFL
Ca in 5TD4
Ca in 5TF9
Ca in 5TAK
Ca in 5TAE
Ca in 5TAJ
Ca in 5TAI
Ca in 5T9T
Ca in 5TAD