Mercury »
PDB 5lf8-6bvg »
5xfe »
Mercury in PDB 5xfe: Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns)
Protein crystallography data
The structure of Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns), PDB code: 5xfe
was solved by
K.Yamashita,
D.Pan,
T.Okuda,
T.Murai,
A.Kodan,
T.Yamaguchi,
K.Gomi,
N.Kajiyama,
H.Kato,
H.Ago,
M.Yamamoto,
T.Nakatsu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.57 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 47.900, 77.030, 84.530, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.2 / 21.3 |
Other elements in 5xfe:
The structure of Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns) also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Mercury Binding Sites:
The binding sites of Mercury atom in the Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns)
(pdb code 5xfe). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 2 binding sites of Mercury where determined in the Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns), PDB code: 5xfe:
Jump to Mercury binding site number: 1; 2;
In total 2 binding sites of Mercury where determined in the Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns), PDB code: 5xfe:
Jump to Mercury binding site number: 1; 2;
Mercury binding site 1 out of 2 in 5xfe
Go back to
Mercury binding site 1 out
of 2 in the Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns)
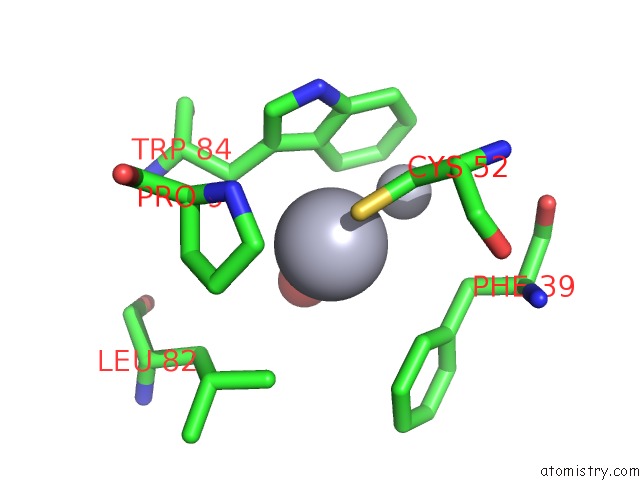
Mono view
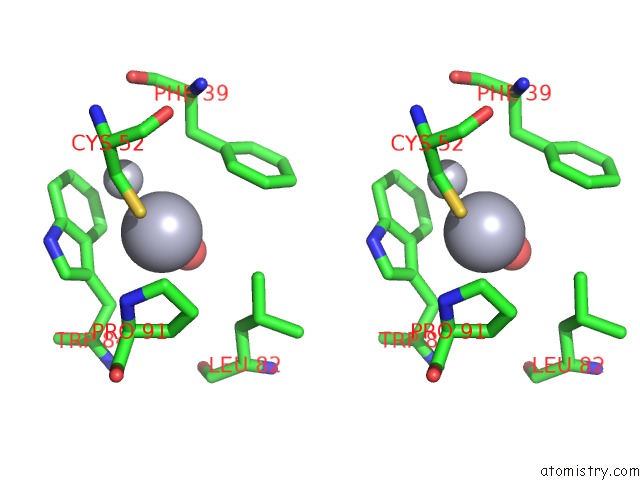
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns) within 5.0Å range:
|
Mercury binding site 2 out of 2 in 5xfe
Go back to
Mercury binding site 2 out
of 2 in the Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns)
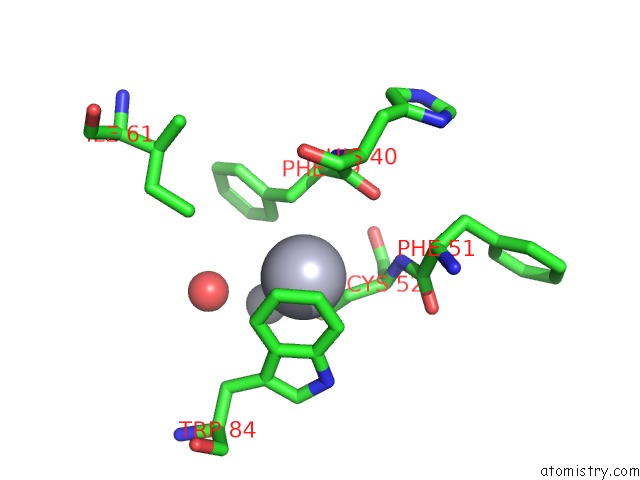
Mono view
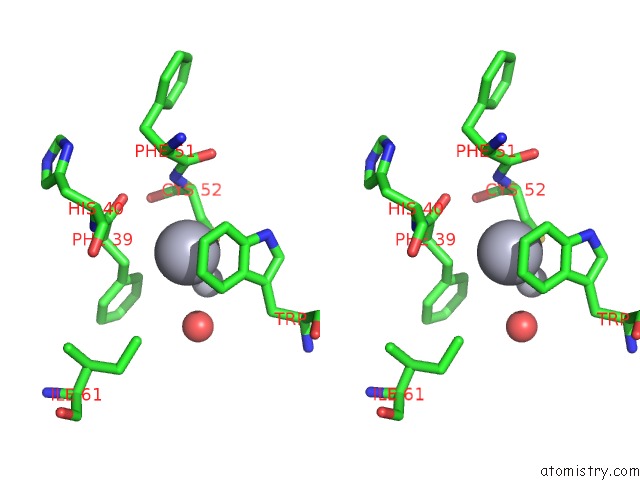
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns) within 5.0Å range:
|
Reference:
K.Yamashita,
N.Kuwabara,
T.Nakane,
T.Murai,
E.Mizohata,
M.Sugahara,
D.Pan,
T.Masuda,
M.Suzuki,
T.Sato,
A.Kodan,
T.Yamaguchi,
E.Nango,
T.Tanaka,
K.Tono,
Y.Joti,
T.Kameshima,
T.Hatsui,
M.Yabashi,
H.Manya,
T.Endo,
R.Kato,
T.Senda,
H.Kato,
S.Iwata,
H.Ago,
M.Yamamoto,
F.Yumoto,
T.Nakatsu.
Experimental Phase Determination with Selenomethionine or Mercury-Derivatization in Serial Femtosecond Crystallography Iucrj V. 4 639 2017.
ISSN: ESSN 2052-2525
PubMed: 28989719
DOI: 10.1107/S2052252517008557
Page generated: Fri Aug 8 11:01:24 2025
ISSN: ESSN 2052-2525
PubMed: 28989719
DOI: 10.1107/S2052252517008557
Last articles
K in 7QQVK in 7QQT
K in 7QQR
K in 7QQS
K in 7QQQ
K in 7QQP
K in 7QQO
K in 7QK5
K in 7QIX
K in 7QNO