Mercury »
PDB 6rkn-7byf »
6sac »
Mercury in PDB 6sac: N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment
Enzymatic activity of N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment
All present enzymatic activity of N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment:
4.2.1.1;
4.2.1.1;
Protein crystallography data
The structure of N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment, PDB code: 6sac
was solved by
S.Gloeckner,
A.Heine,
G.Klebe,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.95 / 1.02 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.374, 41.286, 72.281, 90.00, 104.82, 90.00 |
| R / Rfree (%) | 12.6 / 14.2 |
Other elements in 6sac:
The structure of N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
| Sodium | (Na) | 2 atoms |
Mercury Binding Sites:
The binding sites of Mercury atom in the N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment
(pdb code 6sac). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 2 binding sites of Mercury where determined in the N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment, PDB code: 6sac:
Jump to Mercury binding site number: 1; 2;
In total 2 binding sites of Mercury where determined in the N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment, PDB code: 6sac:
Jump to Mercury binding site number: 1; 2;
Mercury binding site 1 out of 2 in 6sac
Go back to
Mercury binding site 1 out
of 2 in the N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment
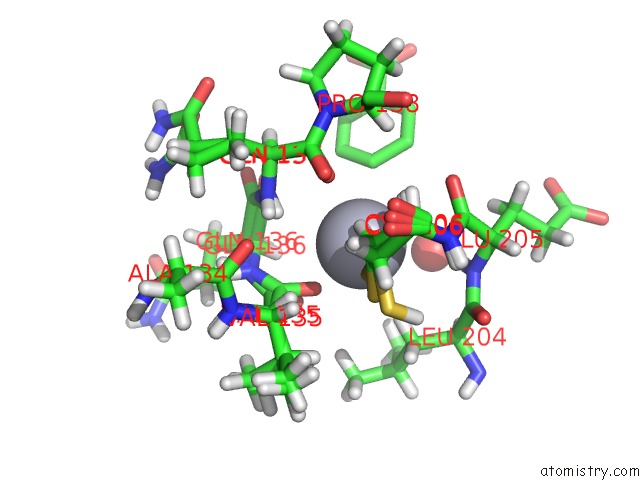
Mono view
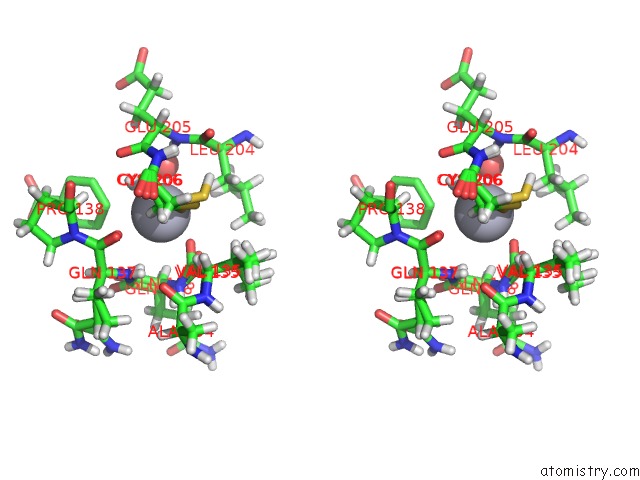
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment within 5.0Å range:
|
Mercury binding site 2 out of 2 in 6sac
Go back to
Mercury binding site 2 out
of 2 in the N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment
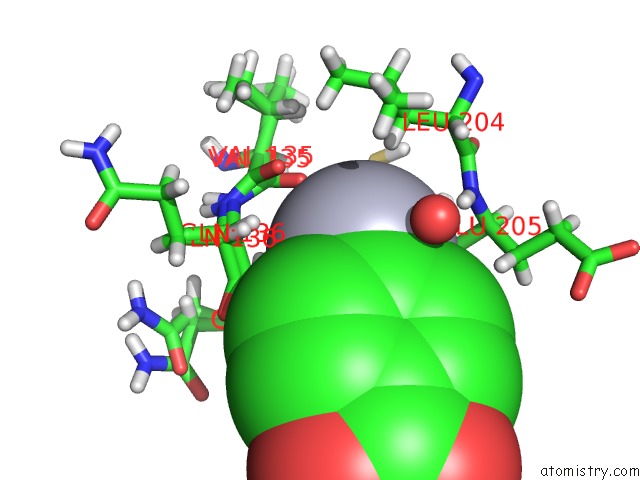
Mono view
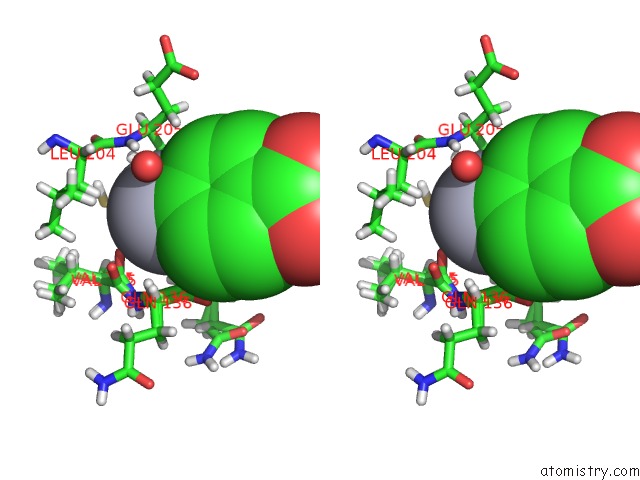
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of N-Terminal Expression Tag Remainder of Human Carbonic Anhydrase II Covalently Modified By Fragment within 5.0Å range:
|
Reference:
S.Glockner,
A.Heine,
G.Klebe.
A Proof-of-Concept Fragment Screening of A Hit-Validated 96-Compounds Library Against Human Carbonic Anhydrase II. Biomolecules V. 10 2020.
ISSN: ESSN 2218-273X
PubMed: 32235320
DOI: 10.3390/BIOM10040518
Page generated: Sun Aug 11 08:09:55 2024
ISSN: ESSN 2218-273X
PubMed: 32235320
DOI: 10.3390/BIOM10040518
Last articles
F in 7NVMF in 7NW2
F in 7NW0
F in 7NVL
F in 7NVX
F in 7NVV
F in 7NVO
F in 7NTH
F in 7NTI
F in 7NPC