Mercury »
PDB 5lf8-6bvg »
5n83 »
Mercury in PDB 5n83: Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative
Protein crystallography data
The structure of Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative, PDB code: 5n83
was solved by
A.K.Singh,
M.J.Van Raaij,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 82.14 / 2.76 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.116, 164.285, 96.432, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.3 / 24.8 |
Mercury Binding Sites:
The binding sites of Mercury atom in the Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative
(pdb code 5n83). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 5 binding sites of Mercury where determined in the Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative, PDB code: 5n83:
Jump to Mercury binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Mercury where determined in the Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative, PDB code: 5n83:
Jump to Mercury binding site number: 1; 2; 3; 4; 5;
Mercury binding site 1 out of 5 in 5n83
Go back to
Mercury binding site 1 out
of 5 in the Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative
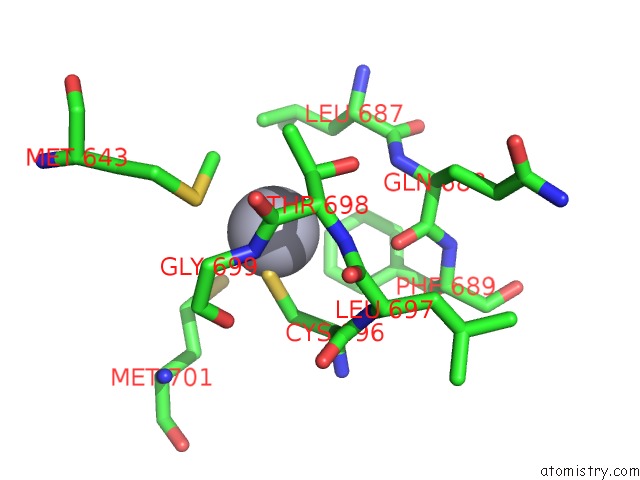
Mono view
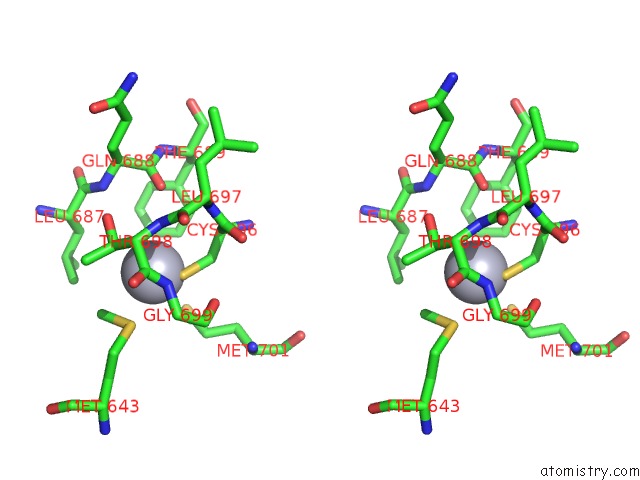
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative within 5.0Å range:
|
Mercury binding site 2 out of 5 in 5n83
Go back to
Mercury binding site 2 out
of 5 in the Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative
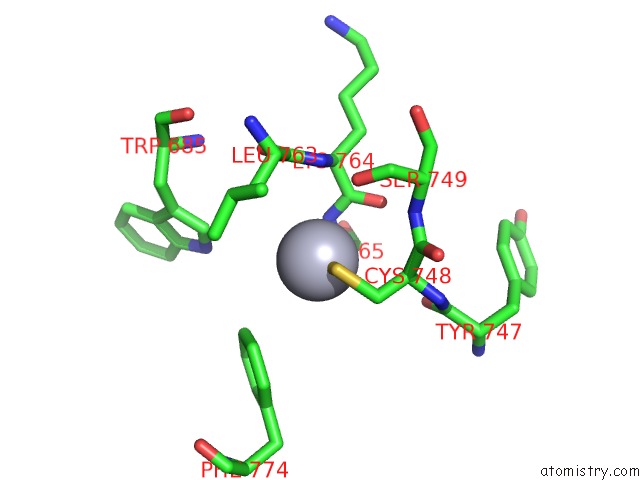
Mono view
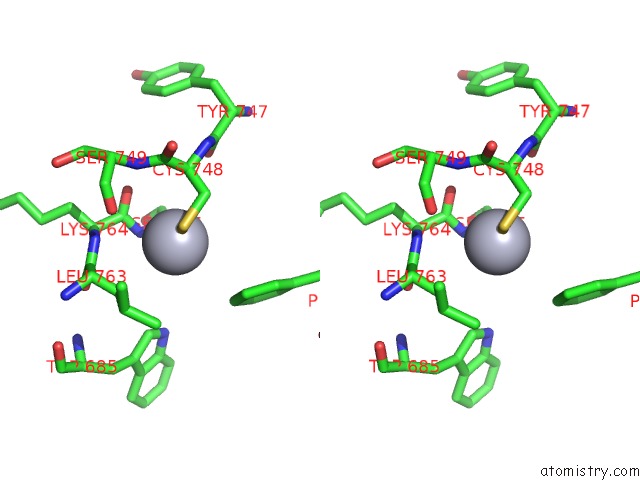
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative within 5.0Å range:
|
Mercury binding site 3 out of 5 in 5n83
Go back to
Mercury binding site 3 out
of 5 in the Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative
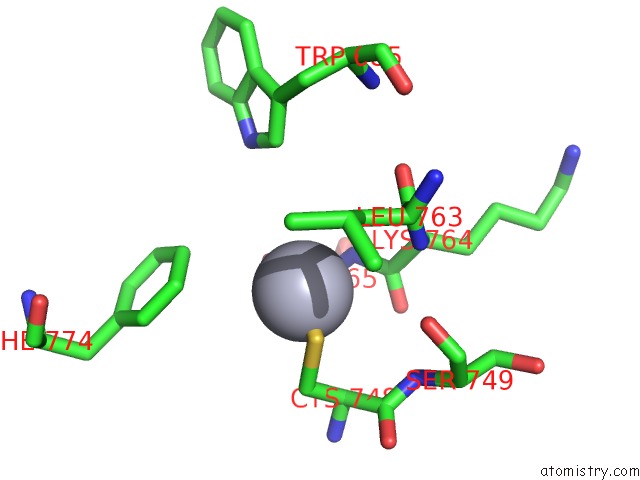
Mono view
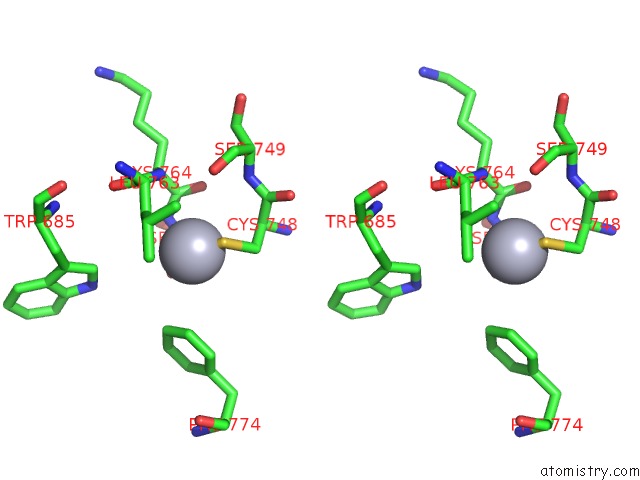
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 3 of Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative within 5.0Å range:
|
Mercury binding site 4 out of 5 in 5n83
Go back to
Mercury binding site 4 out
of 5 in the Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative
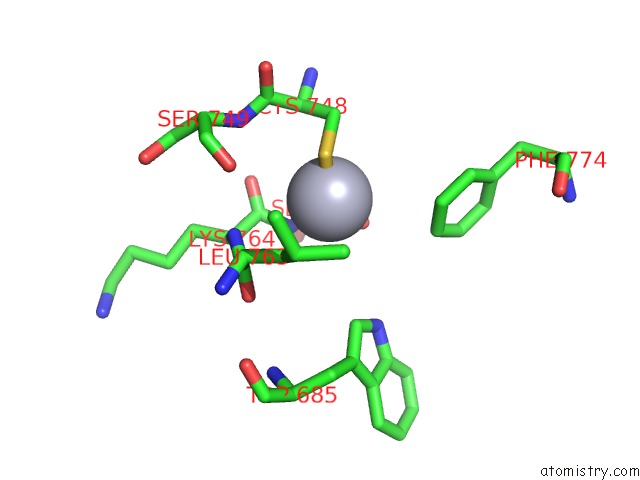
Mono view
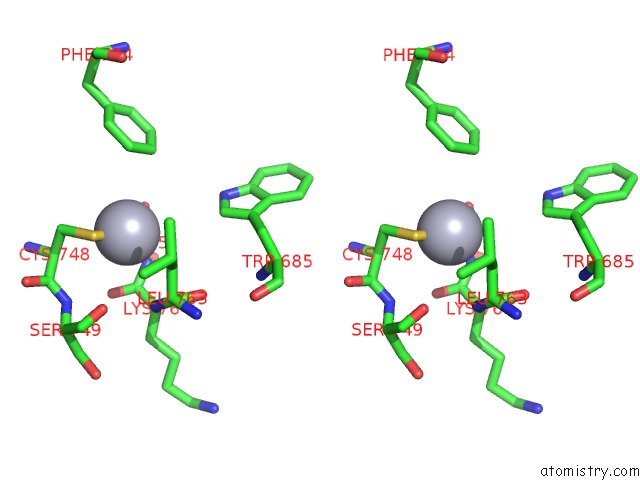
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 4 of Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative within 5.0Å range:
|
Mercury binding site 5 out of 5 in 5n83
Go back to
Mercury binding site 5 out
of 5 in the Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative
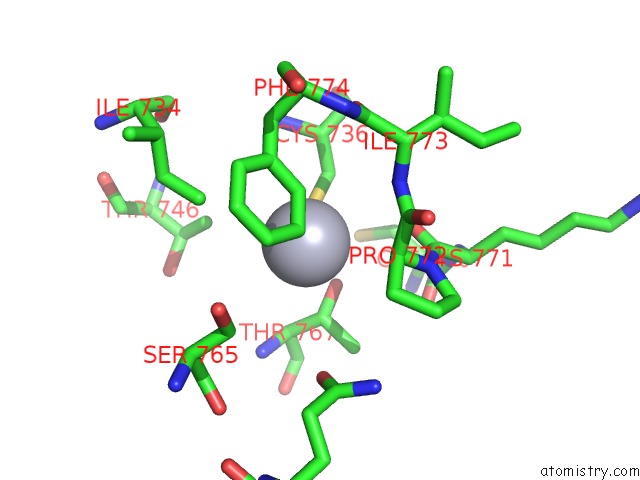
Mono view
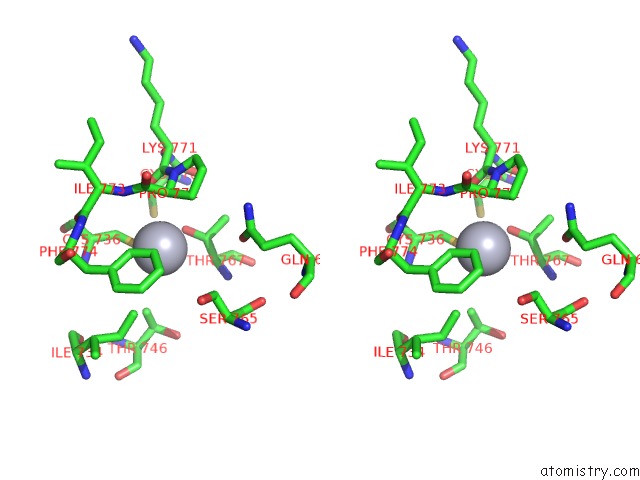
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 5 of Structure of the Distal Domain of Mouse Adenovirus 2 Fibre, Methylmercury Chloride Derivative within 5.0Å range:
|
Reference:
A.K.Singh,
T.H.Nguyen,
M.Z.Vidovszky,
B.Harrach,
M.Benko,
A.Kirwan,
L.Joshi,
M.Kilcoyne,
M.A.Berbis,
F.J.Canada,
J.Jimenez-Barbero,
M.Menendez,
S.S.Wilson,
B.A.Bromme,
J.G.Smith,
M.J.Van Raaij.
Structure and N-Acetylglucosamine Binding of the Distal Domain of Mouse Adenovirus 2 Fibre. J. Gen. Virol. V. 99 1494 2018.
ISSN: ESSN 1465-2099
PubMed: 30277856
DOI: 10.1099/JGV.0.001145
Page generated: Fri Aug 8 10:57:00 2025
ISSN: ESSN 1465-2099
PubMed: 30277856
DOI: 10.1099/JGV.0.001145
Last articles
I in 8D22I in 8CXW
I in 8D00
I in 8CT8
I in 8CUF
I in 8CJ7
I in 8CNZ
I in 8C3P
I in 8CEV
I in 8C7R