Mercury »
PDB 5lf8-6bvg »
6brs »
Mercury in PDB 6brs: The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset
Protein crystallography data
The structure of The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset, PDB code: 6brs
was solved by
R.Grinter,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.65 / 2.30 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.531, 127.369, 132.200, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.9 / 24.2 |
Mercury Binding Sites:
The binding sites of Mercury atom in the The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset
(pdb code 6brs). This binding sites where shown within
5.0 Angstroms radius around Mercury atom.
In total 5 binding sites of Mercury where determined in the The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset, PDB code: 6brs:
Jump to Mercury binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Mercury where determined in the The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset, PDB code: 6brs:
Jump to Mercury binding site number: 1; 2; 3; 4; 5;
Mercury binding site 1 out of 5 in 6brs
Go back to
Mercury binding site 1 out
of 5 in the The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset
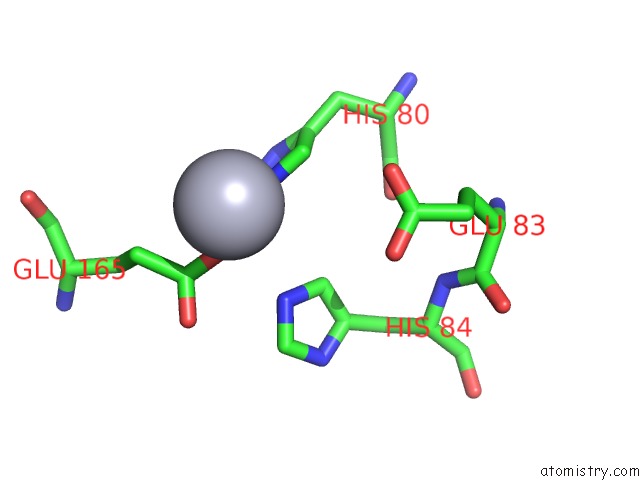
Mono view
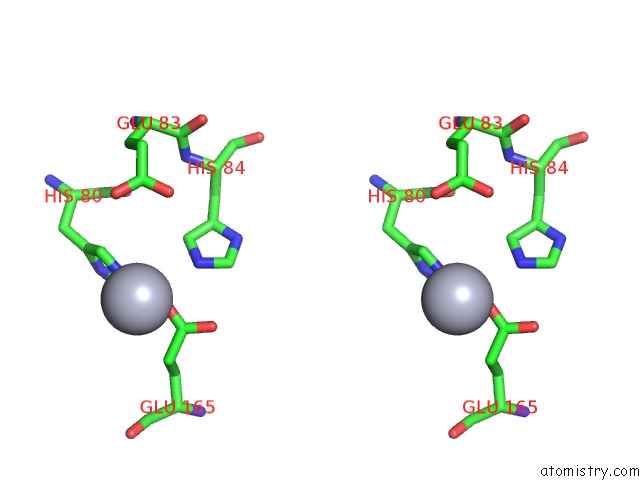
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 1 of The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset within 5.0Å range:
|
Mercury binding site 2 out of 5 in 6brs
Go back to
Mercury binding site 2 out
of 5 in the The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset
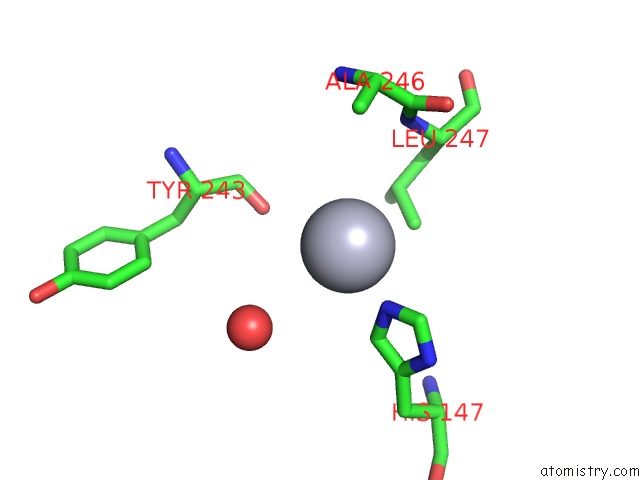
Mono view
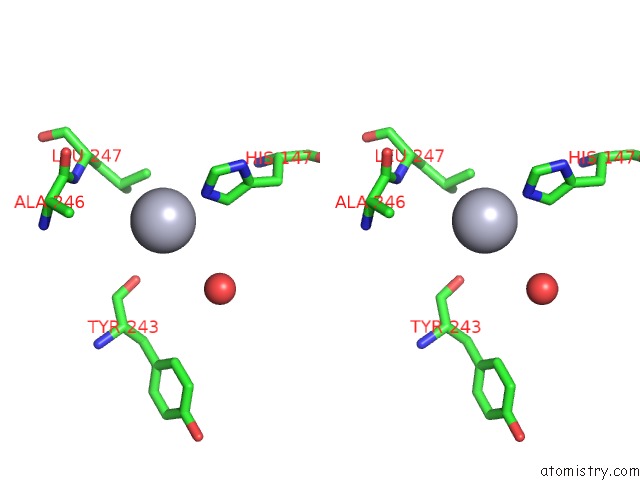
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 2 of The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset within 5.0Å range:
|
Mercury binding site 3 out of 5 in 6brs
Go back to
Mercury binding site 3 out
of 5 in the The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset
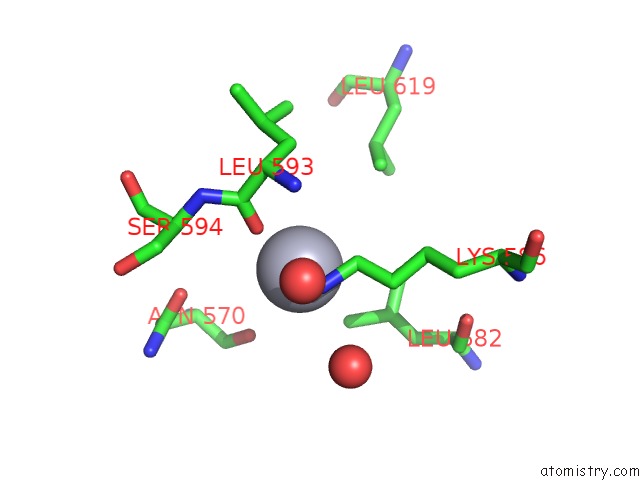
Mono view
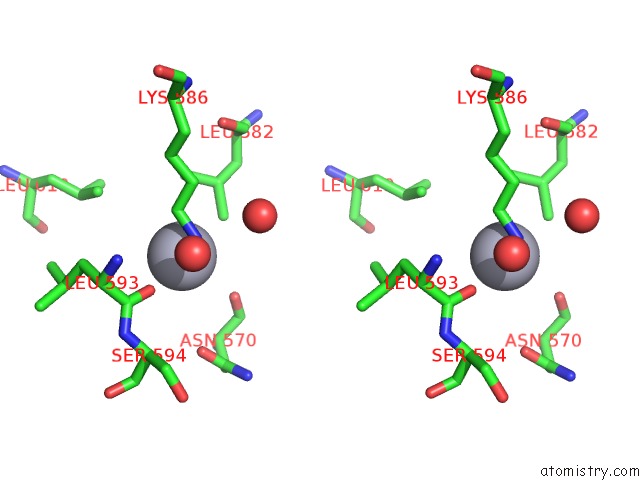
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 3 of The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset within 5.0Å range:
|
Mercury binding site 4 out of 5 in 6brs
Go back to
Mercury binding site 4 out
of 5 in the The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset
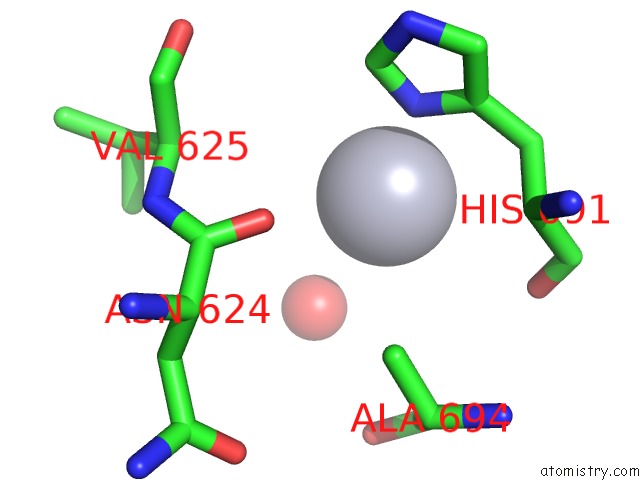
Mono view
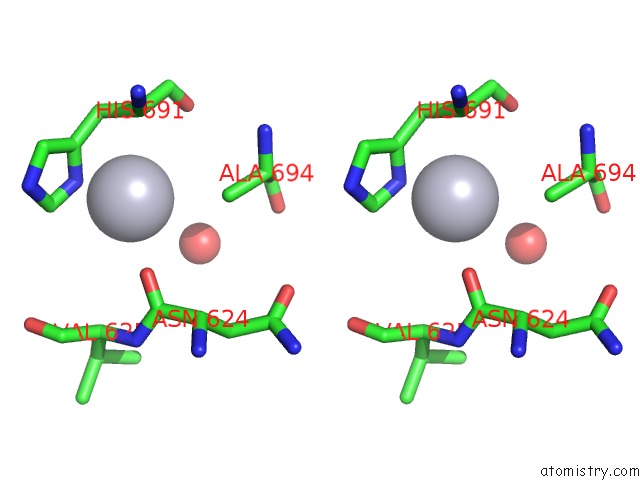
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 4 of The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset within 5.0Å range:
|
Mercury binding site 5 out of 5 in 6brs
Go back to
Mercury binding site 5 out
of 5 in the The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset
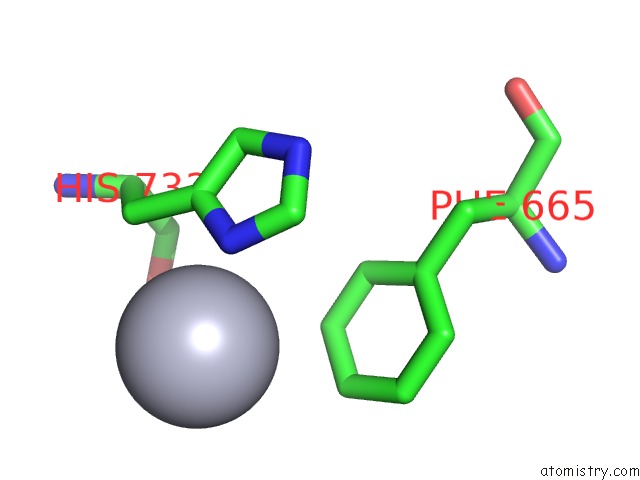
Mono view
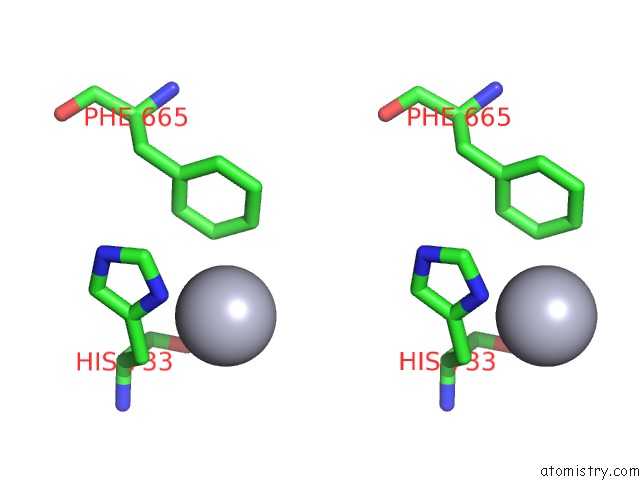
Stereo pair view

Mono view

Stereo pair view
A full contact list of Mercury with other atoms in the Hg binding
site number 5 of The Crystal Structure of the Ferredoxin Protease Fusc in Complex with Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset within 5.0Å range:
|
Reference:
R.Grinter,
I.D.Hay,
J.Song,
J.Wang,
D.Teng,
V.Dhanesakaran,
J.J.Wilksch,
M.R.Davies,
D.Littler,
S.A.Beckham,
I.R.Henderson,
R.A.Strugnell,
G.Dougan,
T.Lithgow.
Fusc, A Member of the M16 Protease Family Acquired By Bacteria For Iron Piracy Against Plants. Plos Biol. V. 16 06026 2018.
ISSN: ESSN 1545-7885
PubMed: 30071011
DOI: 10.1371/JOURNAL.PBIO.2006026
Page generated: Fri Aug 8 11:02:35 2025
ISSN: ESSN 1545-7885
PubMed: 30071011
DOI: 10.1371/JOURNAL.PBIO.2006026
Last articles
Mg in 1W55Mg in 1W54
Mg in 1W4B
Mg in 1W49
Mg in 1W46
Mg in 1W2Y
Mg in 1VQN
Mg in 1W25
Mg in 1W23
Mg in 1W1Z